class: center, middle, inverse, title-slide .title[ # Data vizualisation with <strong>R</strong> (session 4) ] .subtitle[ ## M2 Statistics and Econometrics ] .author[ ### <a href="http://www.thibault.laurent.free.fr">Thibault Laurent</a> ] .institute[ ### Toulouse School of Economics, CNRS ] .date[ ### Last update: 2023-10-04 ] --- class: inverse, middle, center background-image: url(https://cran.r-project.org/Rlogo.svg) background-size: contain # Table of contents #### 1. Nice tools for writting a report #### 2. Base graphics VS **ggplot2** #### 3. Original statistical graphics #### 4. Interactive graphics --- class: inverse, middle, center background-image: url(https://cran.r-project.org/Rlogo.svg) background-size: contain # Before starting --- background-image: url(https://cran.r-project.org/Rlogo.svg) background-size: 100px background-position: 90% 8% # Packages and software versions This document has been compiled under this version: R version 4.3.1 (2023-06-16). Install the following packages and load them: ```r install.packages(c("broom", "gapminder", "ggcorrplot", "ggiraph", "ggpol", "ggridges", "ggtern", "kableExtra", "knitr", "plotly", "RColorBrewer", "shiny", "stargazer", "survival", "survminer", "visreg", "vcd"), dependencies = TRUE) ``` ```r require("gapminder") # use data from https://www.gapminder.org/ require("ggcorrplot") # coorelation matrix with ggplot2 style require("ggiraph") # interactive plot require("ggridges") # multi density plot require("ggpol") # ggplot2 extensions require("ggtern") # ternary diagram require("kableExtra") # include nice Table in Markdown document require("plotly") # interactive graphics require("RColorBrewer") # color palettes require("stargazer") # inlude latex/html regression table require("ggplot2") # data visualization require("visreg") # visualization of regression models require("shiny") # shiny App require("survival") # survival data analysis require("survminer") # visualization of survival data analysis require("vcd") # visualization of categorical data ``` --- class: inverse, middle, center background-image: url(https://cran.r-project.org/Rlogo.svg) background-size: contain # 1. Nice tools for writting a report --- # Include tables in a **R** Markdown document * **R** Markdown is very useful for writing reports which contain text, codes and results of codes. * Use function `kable()` in `knitr` package permits to print a table ````markdown ```{r, results = 'asis'} knitr::kable(head(iris[1:5, ])) ``` ```` ```r kableExtra::kbl(head(iris[1:5, ])) ``` <table> <thead> <tr> <th style="text-align:right;"> Sepal.Length </th> <th style="text-align:right;"> Sepal.Width </th> <th style="text-align:right;"> Petal.Length </th> <th style="text-align:right;"> Petal.Width </th> <th style="text-align:left;"> Species </th> </tr> </thead> <tbody> <tr> <td style="text-align:right;"> 5.1 </td> <td style="text-align:right;"> 3.5 </td> <td style="text-align:right;"> 1.4 </td> <td style="text-align:right;"> 0.2 </td> <td style="text-align:left;"> setosa </td> </tr> <tr> <td style="text-align:right;"> 4.9 </td> <td style="text-align:right;"> 3.0 </td> <td style="text-align:right;"> 1.4 </td> <td style="text-align:right;"> 0.2 </td> <td style="text-align:left;"> setosa </td> </tr> <tr> <td style="text-align:right;"> 4.7 </td> <td style="text-align:right;"> 3.2 </td> <td style="text-align:right;"> 1.3 </td> <td style="text-align:right;"> 0.2 </td> <td style="text-align:left;"> setosa </td> </tr> <tr> <td style="text-align:right;"> 4.6 </td> <td style="text-align:right;"> 3.1 </td> <td style="text-align:right;"> 1.5 </td> <td style="text-align:right;"> 0.2 </td> <td style="text-align:left;"> setosa </td> </tr> <tr> <td style="text-align:right;"> 5.0 </td> <td style="text-align:right;"> 3.6 </td> <td style="text-align:right;"> 1.4 </td> <td style="text-align:right;"> 0.2 </td> <td style="text-align:left;"> setosa </td> </tr> </tbody> </table> --- # Another example from **kableExtra** ```r vs_dt <- iris[1:5, ] vs_dt[1:4] <- lapply(vs_dt[1:4], function(x) { cell_spec(x, bold = T, color = spec_color(x, end = 0.9), font_size = spec_font_size(x)) }) vs_dt[5] <- cell_spec(vs_dt[[5]], color = "white", bold = T, background = spec_color(1:5, end = 0.9, option = "A", direction = -1)) kbl(vs_dt, escape = F, align = "c") %>% kable_classic("striped", full_width = F) ``` <table class=" lightable-classic lightable-striped" style='font-family: "Arial Narrow", "Source Sans Pro", sans-serif; width: auto !important; margin-left: auto; margin-right: auto;'> <thead> <tr> <th style="text-align:center;"> Sepal.Length </th> <th style="text-align:center;"> Sepal.Width </th> <th style="text-align:center;"> Petal.Length </th> <th style="text-align:center;"> Petal.Width </th> <th style="text-align:center;"> Species </th> </tr> </thead> <tbody> <tr> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(187, 223, 39, 1) !important;font-size: 16px;">5.1</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(92, 200, 99, 1) !important;font-size: 15px;">3.5</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(37, 131, 142, 1) !important;font-size: 12px;">1.4</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(37, 131, 142, 1) !important;font-size: 12px;">0.2</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: white !important;border-radius: 4px; padding-right: 4px; padding-left: 4px; background-color: rgba(254, 206, 145, 1) !important;">setosa</span> </td> </tr> <tr> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(31, 154, 138, 1) !important;font-size: 13px;">4.9</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(68, 1, 84, 1) !important;font-size: 8px;">3</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(37, 131, 142, 1) !important;font-size: 12px;">1.4</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(37, 131, 142, 1) !important;font-size: 12px;">0.2</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: white !important;border-radius: 4px; padding-right: 4px; padding-left: 4px; background-color: rgba(242, 100, 92, 1) !important;">setosa</span> </td> </tr> <tr> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(67, 62, 133, 1) !important;font-size: 10px;">4.7</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(53, 96, 141, 1) !important;font-size: 11px;">3.2</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(68, 1, 84, 1) !important;font-size: 8px;">1.3</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(37, 131, 142, 1) !important;font-size: 12px;">0.2</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: white !important;border-radius: 4px; padding-right: 4px; padding-left: 4px; background-color: rgba(161, 48, 126, 1) !important;">setosa</span> </td> </tr> <tr> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(68, 1, 84, 1) !important;font-size: 8px;">4.6</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(69, 53, 129, 1) !important;font-size: 9px;">3.1</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(187, 223, 39, 1) !important;font-size: 16px;">1.5</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(37, 131, 142, 1) !important;font-size: 12px;">0.2</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: white !important;border-radius: 4px; padding-right: 4px; padding-left: 4px; background-color: rgba(70, 16, 120, 1) !important;">setosa</span> </td> </tr> <tr> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(77, 195, 107, 1) !important;font-size: 14px;">5</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(187, 223, 39, 1) !important;font-size: 16px;">3.6</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(37, 131, 142, 1) !important;font-size: 12px;">1.4</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: rgba(37, 131, 142, 1) !important;font-size: 12px;">0.2</span> </td> <td style="text-align:center;"> <span style=" font-weight: bold; color: white !important;border-radius: 4px; padding-right: 4px; padding-left: 4px; background-color: rgba(0, 0, 4, 1) !important;">setosa</span> </td> </tr> </tbody> </table> More informations [here](https://cran.r-project.org/web/packages/kableExtra/vignettes/awesome_table_in_html.html). --- # Print the result of a linear regression model One can use function `kbl()` after applying function `tidy()` on the `lm` class of object ```r lm(Sepal.Length ~ Species, data = iris) %>% broom::tidy() %>% kbl() ``` <table> <thead> <tr> <th style="text-align:left;"> term </th> <th style="text-align:right;"> estimate </th> <th style="text-align:right;"> std.error </th> <th style="text-align:right;"> statistic </th> <th style="text-align:right;"> p.value </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> (Intercept) </td> <td style="text-align:right;"> 5.006 </td> <td style="text-align:right;"> 0.0728022 </td> <td style="text-align:right;"> 68.761639 </td> <td style="text-align:right;"> 0 </td> </tr> <tr> <td style="text-align:left;"> Speciesversicolor </td> <td style="text-align:right;"> 0.930 </td> <td style="text-align:right;"> 0.1029579 </td> <td style="text-align:right;"> 9.032819 </td> <td style="text-align:right;"> 0 </td> </tr> <tr> <td style="text-align:left;"> Speciesvirginica </td> <td style="text-align:right;"> 1.582 </td> <td style="text-align:right;"> 0.1029579 </td> <td style="text-align:right;"> 15.365506 </td> <td style="text-align:right;"> 0 </td> </tr> </tbody> </table> More informations [here](https://zief0002.github.io/book-8252/pretty-printing-tables-in-markdown.html). --- # Compare different regression models .pull-left[ `stargazer()` from `stargazer` permits to print several regression table ```r mod_1 <- lm(Sepal.Length ~ Petal.Length, data = iris) mod_2 <- lm(Sepal.Length ~ Petal.Width, data = iris) stargazer::stargazer(mod_1, mod_2, type = "html", title = "Regression results", header = F) ``` ] .pull-right[ <font size="1"> <table style="text-align:center"><caption><strong>Regression results</strong></caption> <tr><td colspan="3" style="border-bottom: 1px solid black"></td></tr><tr><td style="text-align:left"></td><td colspan="2"><em>Dependent variable:</em></td></tr> <tr><td></td><td colspan="2" style="border-bottom: 1px solid black"></td></tr> <tr><td style="text-align:left"></td><td colspan="2">Sepal.Length</td></tr> <tr><td style="text-align:left"></td><td>(1)</td><td>(2)</td></tr> <tr><td colspan="3" style="border-bottom: 1px solid black"></td></tr><tr><td style="text-align:left">Petal.Length</td><td>0.409<sup>***</sup></td><td></td></tr> <tr><td style="text-align:left"></td><td>(0.019)</td><td></td></tr> <tr><td style="text-align:left"></td><td></td><td></td></tr> <tr><td style="text-align:left">Petal.Width</td><td></td><td>0.889<sup>***</sup></td></tr> <tr><td style="text-align:left"></td><td></td><td>(0.051)</td></tr> <tr><td style="text-align:left"></td><td></td><td></td></tr> <tr><td style="text-align:left">Constant</td><td>4.307<sup>***</sup></td><td>4.778<sup>***</sup></td></tr> <tr><td style="text-align:left"></td><td>(0.078)</td><td>(0.073)</td></tr> <tr><td style="text-align:left"></td><td></td><td></td></tr> <tr><td colspan="3" style="border-bottom: 1px solid black"></td></tr><tr><td style="text-align:left">Observations</td><td>150</td><td>150</td></tr> <tr><td style="text-align:left">R<sup>2</sup></td><td>0.760</td><td>0.669</td></tr> <tr><td style="text-align:left">Adjusted R<sup>2</sup></td><td>0.758</td><td>0.667</td></tr> <tr><td style="text-align:left">Residual Std. Error (df = 148)</td><td>0.407</td><td>0.478</td></tr> <tr><td style="text-align:left">F Statistic (df = 1; 148)</td><td>468.550<sup>***</sup></td><td>299.167<sup>***</sup></td></tr> <tr><td colspan="3" style="border-bottom: 1px solid black"></td></tr><tr><td style="text-align:left"><em>Note:</em></td><td colspan="2" style="text-align:right"><sup>*</sup>p<0.1; <sup>**</sup>p<0.05; <sup>***</sup>p<0.01</td></tr> </table> ] --- # 👩🎓 Training ### Exercise 4.1 * Insert in a Markdown document the correlation matrix of the numeric variables of the `iris` data. * Insert the table of results of a regression analysis of the `iris` data --- class: inverse, middle, center background-image: url(https://cran.r-project.org/Rlogo.svg) background-size: contain # 2. Base graphics VS **ggplot2** --- # Base graphics VS **ggplot2** There are two main approaches for doing graphics I. Base functions programming. II. Use package `ggplot2` which uses its own syntax. Comparisons of the two approaches: * Most of the time, the two approaches are not complementary. * Objective of this section: obtain the same graphics by using the two approaches. * Some references: + [Comparing ggplot2 and R Base Graphics](https://flowingdata.com/2016/03/22/comparing-ggplot2-and-r-base-graphics/) + [Graphics with R (in French)](http://www.thibault.laurent.free.fr/cours/R_intro/chapitre_4.html) + [Univariate Graphs](https://rkabacoff.github.io/datavis/Univariate.html) * Package `esquisse` allows to obtain `ggplot2` codes using an interface **Remark:** actually, there is a third approach by using `lattice` package (see [this link](https://www.londonr.org/wp-content/uploads/sites/2/presentations/LondonR_-_lattice_vs_ggplot2_-_Richard_Pugh_and_Andy_Nicholls_-_20130910.pdf) for more informations), but we will not present this solution --- # Base graphics syntax 1. (optionnal): use `par()` function for defining the margin, the size of the labels, etc. 2. Open a device with a first-level plot function like `plot()`, `hist()`, `barplot()`, `boxplot()`, etc. 3. Customize the plot by using functions like `points()`, `lines()`, `text()`, `legend()` ```r op <- par(oma = c(1, 1, 0, 1), las = 1) boxplot(Sepal.Length ~ Species, data = iris) points(as.numeric(iris$Species) + rnorm(150, 0, 0.1), iris$Sepal.Length) points(c(1, 2, 3), tapply(iris$Sepal.Length, iris$Species, mean), col = "red", pch = 16, cex = 2) ``` <!-- --> ```r par(op) ``` --- # **ggplot2** syntax 1. use `ggplot()` function for specifying: + data frame containing the data to be plotted + the mapping of the variables to visual properties of the graph within the `aes()` function. 2. Add the operator `+` followed by the geometric objects `Geoms` (points, lines, bars, etc.) that can be placed on a graph using functions that look `geom_xxx()` 3. Add the operator `+` followed by the `scales` control which define how variables are mapped to the visual characteristics of the plot. Use functions which look `scale_xxx()` 4. Add the operator `+` followed by the `facets` which reproduce a graph for each level of a given variable. Use functions that look `facet_xxx()` .pull-left[ ```r data("diamonds") ggplot(diamonds, aes(x = carat, y = price)) + geom_point() + ggtitle("My scatter plot") ``` ] .pull-right[ <!-- --> ] --- # Barplot with **ggplot2** ```r ggplot(diamonds) + aes(x = cut) + geom_bar(stat = "count") + xlab("quality degree") + ylab("Size") + ggtitle("Quality diamonds") ``` <!-- --> --- # Barplot with graphic base .pull-left[ We first need to define the contingency table (`table` object) ```r tab_cut <- table(diamonds$cut) ``` We then use `barplot()` function and `abline()` to get the horizontal lines like with `ggplot2` ```r par(las = 1) barplot(tab_cut, col = "#AFC0CB", border = FALSE, main = "Quality diamonds", xlab = "quality degree", ylab = "size", cex.axis = 0.8) abline(h = seq(0, 20000, by = 2500), col = "lightgray", lty = "dotted") ``` ] .pull-right[ <!-- --> ] --- # Plotting a time serie We create a vector of numeric values: ```r serie <- c(161.31, 154.00, 161.94, 160.23, 173.20, 170.21, 163.97, 161.70, 144.91, 145.31, 140.50, 139.58, 135.60, 124.40, 132.24, 150.51, 146.56, 153.00, 151.78, 160.65, 158.32, 158.06, 153.50, 161.95, 167.00, 175.00, 180.48, 173.82, 160.05, 152.80, 153.58, 145.00, 142.98, 145.35) ``` We create a vector of type `Date`: ```r date_serie <- seq(as.Date("2015/1/1"), by = "month", length.out = 34) ``` For using `ggplot2`, user needs to create a `data.frame` (or `tibble`) object (which is not the case with the **R** base code which allows the use of vectors in most of the functions): ```r serie_df <- data.frame(date_serie = date_serie, serie = serie) ``` --- # Plot a time serie with **ggplot2** ```r ggplot(serie_df) + aes(x = date_serie, y = serie) + geom_line(linetype = 2, colour = "blue") + xlab("Months") + ylab("Index") + ggtitle("Evolution") ``` <!-- --> --- # Plot a time serie with **R** base code ```r par(las = 1) plot(serie ~ date_serie, data = serie_df, type = "l", col = "royalblue", lty = 2, main = "Evolution", xlab = "Months", ylab = "Index", cex.axis = 0.8) abline(h = seq(130, 180, by = 10), v = date_serie[seq(1, 32, 6)], col = "lightgray", lty = "dotted") ``` <!-- --> --- # Histogram and density plot with **ggplot2** ```r ggplot(diamonds) + aes(x = price) + geom_histogram(aes(y = after_stat(density)), fill = "blue", colour = "black", bins = 30) + geom_density(colour = "red", adjust = 2) + stat_function(fun = dnorm, args = c(mean = mean(diamonds$price), sd = sd(diamonds$price))) + xlab("Price") + ggtitle("Distribution price") ``` <!-- --> --- # Histogram and density plot with with **R** base code ```r par(las = 1, cex.axis = 0.8, cex.lab = 0.8) hist(diamonds$price, freq = F, col = "lightblue", nclass = 30, xlab = "Price", main = "Distribution price") lines(density(diamonds$price),col = "red") x_seq <- seq(-1000, 20000, by = 100) lines(x_seq, dnorm(x_seq, mean(diamonds$price), sd(diamonds$price))) abline(h = seq(0, 0.0005, by = 0.00005), v = seq(0, 20000, by = 2500), col = "lightgray", lty = "dotted") ``` <!-- --> --- # Scatter plot with **ggplot2** We select first a sample of the observations: ```r set.seed(123) # fix a seed diam_ech <- diamonds[sample(nrow(diamonds), 5000, replace = F), ] ggplot(diam_ech) + aes(x = carat, y = price) + geom_point() + geom_smooth(method = "loess") + # Non parametric regression geom_smooth(method = "lm", col = "red") + # Linear regression xlab("Carat") + ylab("Price") + ggtitle("Relationship between Y and X") ``` <!-- --> --- # Scatter plot with **R** base code ```r par(las = 1, cex.axis = 0.8, cex.lab = 0.8) plot(price ~ carat, data = diam_ech, pch = 16, cex = 0.7, xlab = "carat", ylab= "prix", main = "Scatter plot") abline(lm(price ~ carat, data = diam_ech), col = "red", lwd = 3) # values to predict x_carat <- seq(0, 4.5, 0.01) lines(x_carat, predict(loess(price ~ carat, data = diam_ech), data.frame(carat = x_carat)), col = "blue", lwd = 3) abline(h = seq(0, 20000, by = 5000), v = seq(0, 4, by = 0.5), col = "lightgray", lty = "dotted") ``` <!-- --> --- # Parallel boxplot with **ggplot2** ```r ggplot(diam_ech) + aes(x = color, y = price) + geom_boxplot() ``` <!-- --> --- # Parallel boxplot with **R** base code ```r par(las = 1, cex.axis = 0.8, cex.lab = 0.8) boxplot(price ~ color, data = diam_ech, pch = 16, cex = 0.7, xlab = "carat", ylab= "prix") abline(h = seq(0, 20000, by = 2500), v = seq(0, 5, by = 1), col = "lightgray", lty = "dotted") ``` <!-- --> --- # What is conditionnal graphics? **Objective:** it consists in representing the distributions on sub-populations (for example, a population can be splitted with respect to a qualitative variable). It can be done differently: .pull-left[ Represent all the sub-distributions in the same graphic <!-- --> ] .pull-right[ Repeat the same graphic for each sub-sample (can be called `facets`). <!-- --> ] --- # Conditionnal density plot with **ggplot2** It is very easy to plot conditionnal graphics with **ggplot2**. Note that these 3 rows of code represent actually a lot of **R** base codes. Initial graphical parameters are already chosen. ```r ggplot(diam_ech) + aes(x = price, fill = cut) + geom_density(alpha = 0.5) ``` <!-- --> --- # Conditionnal density plot with **R** base code It can be boring to make conditional grahics with **R** base code but ... it is still possible to make some elegant graphics and maybe easier to change optional parameters. .pull-left[ ```r list_price <- split(diam_ech$price, diam_ech$cut) list_density <- lapply(list_price, density) par(las = 1, cex.axis = 0.8, cex.lab = 0.8) plot(range(unlist(lapply( list_density, function(l) range(l$x)))), range(unlist(lapply(list_density, function(l) range(l$y)))), type = "n", xlab = "price", ylab = "density") col_pal <- c("#F8766D", "#A3A500", "#00BF7D", "#00B0F6", "#E76BF3") dont_print <- mapply(lines, list_density, col = col_pal, lwd = 2) abline(h = seq(0, 4*10^(-4), by = 10^(-4)), v = seq(0, 25000, by = 5000), col = "lightgray", lty = "dotted") legend("topright", legend = names(list_density), col = col_pal, lwd = 2, cex = 0.8) ``` ] .pull-right[ <!-- --> ] --- # Conditionnal scatter plot with **ggplot2** .pull-left[ ```r ggplot(diam_ech) + aes(x = carat, y = price) + geom_point() + geom_smooth(aes(colour = cut)) + theme_bw() + xlab("Carat") + ylab("price (in USD)") + ggtitle("Scatter plot") + scale_colour_brewer(name = "Qualité", labels = c("A--", "A-", "A", "A+", "A++"), palette = "Greens") ``` ] .pull-right[ <!-- --> ] --- # Conditionnal scatter plot with **R** base code .pull-left[ ```r par(las = 1, cex.axis = 0.8, cex.lab = 0.8) plot(price ~ carat, data = diam_ech, pch = 16, cex = 0.7, xlab = "carat", ylab = "price (in USD)", main = "Scatter plot") abline(h = seq(0, 20000, by = 5000), v = seq(0, 4, by = 0.5), col = "lightgray", lty = "dotted") list_df <- split(diam_ech, diam_ech$cut) x_carat <- seq(0, 4.5, 0.01) list_loess <- lapply(list_df, function(obj) predict(loess(price ~ carat, data = obj), data.frame(carat = x_carat))) require("RColorBrewer") col_pal <- brewer.pal(length(list_price), "Greens") dont_print <- mapply(lines, rep(list(x_carat), 5), list_loess, col = col_pal, lwd = 3) legend("topright", legend = c("A--", "A-", "A", "A+", "A++"), col = col_pal, lwd = 2, cex = 0.8) ``` ] .pull-right[ <!-- --> ] --- # Conditionnal parallel boxplot with **ggplot2** ```r ggplot(diam_ech) + aes(x = color, y = price, fill = cut) + geom_boxplot() ``` <!-- --> --- # Conditionnal parallel boxplot with **R** base code .pull-left[ ```r col_pal <- c("#F8766D", "#A3A500", "#00BF7D", "#00B0F6", "#E76BF3") par(las = 1, cex.axis = 0.8, cex.lab = 0.8, xpd = T, mar = par()$mar + c(0, 0, 0, 4)) boxplot(price ~ cut + color, data = diam_ech, xlab = "color", ylab = "price", at = c(1:5, 7:11, 13:17, 19:23, 25:29, 31:35, 37:41), col = rep(col_pal, 7), pch = 16, xaxt = "n") axis(1, at = c(3, 9, 15, 21, 27, 33, 39), labels = c("D", "E", "F", "G", "H", "I", "J")) abline(h = seq(0, 20000, by = 2500), col = "lightgray", lty = "dotted") legend(45, 15000, legend = c("A--", "A-", "A", "A+", "A++"), fill = col_pal) ``` ] .pull-right[ <!-- --> ] --- # Facets with **ggplot2** ```r ggplot(diam_ech) + aes(x = price) + geom_density() + facet_wrap(~ cut) ``` <!-- --> --- # 👩🎓 Training ### Exercise 4.2 * Find the code which allows to obtain with **ggplot2** this figure: ```r op <- par(oma = c(1, 1, 0, 1), las = 1) boxplot(Sepal.Length ~ Species, data = iris) points(as.numeric(iris$Species) + rnorm(150, 0, 0.1), iris$Sepal.Length) points(c(1, 2, 3), tapply(iris$Sepal.Length, iris$Species, mean), col = "red", pch = 16, cex = 2) par(op) ``` * Find the code in **R** base code which allows to obtain this figure: ```r data("diamonds") ggplot(diamonds, aes(x = carat, y = price)) + geom_point() + ggtitle("My scatter plot") ``` --- class: inverse, middle, center background-image: url(https://cran.r-project.org/Rlogo.svg) background-size: contain # 3. Original statistical graphics --- # Violin plots It is the combination of density plot (in blue) and boxplot ```r ggplot(diamonds, aes(x = cut, y = price)) + geom_violin(fill = "cornflowerblue") + geom_boxplot(width = .2, fill = "orange", outlier.color = "orange", outlier.size = 2) + labs(title = "Price dist. by cut") ``` <!-- --> --- # Combining jitter and boxplot It is the combination of boxplot and jitter ```r ggplot(diamonds, aes(x = cut, y = price, fill = cut)) + geom_boxjitter(color = "black", jitter.color = "darkgrey", errorbar.draw = TRUE) + theme_minimal() + theme(legend.position = "none") ``` <!-- --> --- # Ridgeline plot It represents the density plot on sub-sample ```r ggplot(diam_ech) + aes(x = price, y = color, fill = color) + geom_density_ridges() + theme_ridges() + labs("Price by levels color") + theme(legend.position = "none") ``` <!-- --> --- # Comparing the means of sub-sample We first compute in a `data.frame` the mean, the standard deviation, the standard error and the confidence interval of a numeric variable with respect to 2 categorical variables ```r library(dplyr) plotdata <- diamonds %>% group_by(color, cut) %>% summarize(n = n(), mean = mean(price), sd = sd(price), se = sd / sqrt(n), ci = qt(0.975, df = n - 1) * sd / sqrt(n)) ``` --- # Represent error bars (1) ⚠️ we get different results with respect to the choosen criteria. Here the SD: ```r ggplot(plotdata) + aes(x = cut, y = mean, group = color, color = color) + geom_point(size = 3) + geom_line(linewidth = 1) + geom_errorbar(aes(ymin = mean - sd, ymax = mean + sd), width = .1) + labs(title = "standard deviation") ``` <!-- --> --- # Represent error bars (2) ⚠️ we get different results with respect to the choosen criteria. Here the SE: ```r ggplot(plotdata) + aes(x = cut, y = mean, group = color, color = color) + geom_point(size = 3) + geom_line(linewidth = 1) + geom_errorbar(aes(ymin = mean - se, ymax = mean + se), width = .1) + labs(title = "standard error") ``` <!-- --> --- # Represent error bars (3) ⚠️ we get different results with respect to the choosen criteria. Here the CI: ```r ggplot(plotdata) + aes(x = cut, y = mean, group = color, color = color) + geom_point(size = 3) + geom_line(linewidth = 1) + geom_errorbar(aes(ymin = mean - ci, ymax = mean + ci), width = .1) + labs(title = "confidence interval") ``` <!-- --> --- # Cleveland dot chart Compare the 2007 life expectancy for Asian country using the gapminder dataset. .pull-left[ ```r data(gapminder, package = "gapminder") library(dplyr) plotdata <- gapminder %>% filter(continent == "Asia" & year == 2007) ggplot(plotdata) + aes(x = lifeExp, y = reorder(country, lifeExp)) + geom_point() ``` ] .pull-right[ <!-- --> ] --- # Area chart Compare the shares of different companies across the time .pull-left[ ```r time_chart <- data.frame( year = rep(as.Date(c(2000, 2005, 2010)), each =3), market_share = c(20, 50, 30, 30, 50, 20, 50, 30, 20), comp = rep(c("a", "b", "c"), 3) ) ggplot(time_chart) + aes(x = year, y = market_share, fill = comp) + geom_area(color = "black") + labs(title = "Market share", subtitle = "2000 to 2010", x = "Year", y = "percentage", fill = "Company") + scale_fill_brewer(palette = "Set2") + theme_minimal() ``` ] .pull-right[ <!-- --> ] --- # Ternary diagram for compositional data Compare the shares of different companies across the time .pull-left[ ```r library(tidyverse) time_chart_wide <- pivot_wider(time_chart, values_from = market_share, names_from = comp) library("ggtern") ggtern(data = time_chart_wide, mapping = aes(x = a, y = b, z = c)) + geom_point(size = 1.5) ``` ] .pull-right[ <!-- --> ] --- # Correlation plot ```r r <- cor(iris[, 1:4], use = "complete.obs") ggcorrplot(r, hc.order = TRUE, type = "lower", lab = TRUE) ``` <!-- --> --- # Linear regression (1) The `visreg()` function takes (1) the model and (2) the variable of interest and plots the conditional relationship, controlling for the other variables (see [this link](https://journal.r-project.org/archive/2017/RJ-2017-046/RJ-2017-046.pdf) for more informations). Example on a numeric variable: ```r res_lm <- lm(Sepal.Length ~ Sepal.Width + Petal.Width + Species, data = iris) visreg(res_lm, "Sepal.Width", gg = TRUE) ``` <!-- --> --- # Linear regression (2) Example on a categorical variable: ```r visreg(res_lm, "Species", gg = TRUE) ``` <!-- --> --- # Logistic regression Model first: ```r iris$binary <- factor(ifelse(iris$Species == "setosa", 1, 0)) res_glm <- glm(binary ~ Sepal.Length, family = binomial(link = "logit"), data = iris) ``` Plot the probability function with respect to one explanatory variable: ```r visreg(res_glm, "Sepal.Length", gg = TRUE, scale="response") ``` <!-- --> --- # Survival plot Model first: ```r sfit <- survfit(Surv(time, status) ~ sex, data = lung) ``` Plot the survival plot: ```r ggsurvplot(sfit, conf.int = TRUE, pval = TRUE, legend.labs = c("M", "F"), legend.title = "Sex", palette = c("cornflowerblue", "indianred3"),title = "Kaplan-Meier", xlab = "Time (days)") ``` 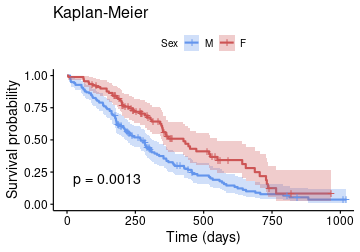<!-- --> --- # Mosaic plot The mosaic plot permits to appreciate the link between two categorical variables: ```r tab <- xtabs(~cut + color, diamonds) mosaic(tab, shade = TRUE, legend = TRUE) ``` 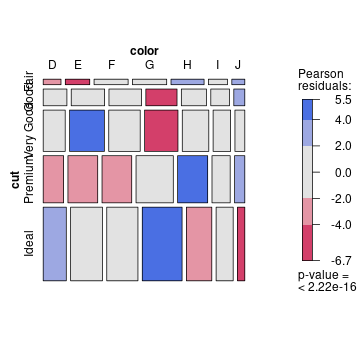<!-- --> --- # 👩🎓 Training ### Exercise 4.3 * On the `lung` data used previously, make a mosaic plot between `status` and `sex` variable. * On the `lung` data, make a ridge plot of variable `age` with respect to `status`. * Make a correlation plot of variables `ph.karno`, `pat.karno`, `meal.cal`, `wt.loss` in the `lung` data. --- class: inverse, middle, center background-image: url(https://cran.r-project.org/Rlogo.svg) background-size: contain # 4. Interactive Graphics --- # **plotly** package (1) * Interactive graphics with respect to library **plotly.js** (in **JavaScript**) * More informations [here](https://plot.ly/r/) * Syntax: + 1st argument: the `data.frame` + argument `x=` gives the name of the `\(x\)` variable; argument `y=` gives the name of the `\(y\)` variable; + argument `color=` gives the name of the conditionnal variable + argument `type=` gives the type of graphic ```r plot_ly(diam_ech, x = ~price, color = ~cut, type = "box") ``` <div class="plotly html-widget html-fill-item-overflow-hidden html-fill-item" id="htmlwidget-843c054f80e4323a248f" style="width:360px;height:288px;"></div> <script type="application/json" data-for="htmlwidget-843c054f80e4323a248f">{"x":{"visdat":{"13c6a2665c8b52":["function () ","plotlyVisDat"]},"cur_data":"13c6a2665c8b52","attrs":{"13c6a2665c8b52":{"x":{},"color":{},"alpha_stroke":1,"sizes":[10,100],"spans":[1,20],"type":"box"}},"layout":{"margin":{"b":40,"l":60,"t":25,"r":10},"xaxis":{"domain":[0,1],"automargin":true,"title":"price"},"yaxis":{"domain":[0,1],"automargin":true},"hovermode":"closest","showlegend":true},"source":"A","config":{"modeBarButtonsToAdd":["hoverclosest","hovercompare"],"showSendToCloud":false},"data":[{"fillcolor":"rgba(253,231,37,0.5)","x":[2397,3300,713,707,987,1053,758,734,5546,1207,403,1116,1441,545,673,5013,1351,3985,829,1656,534,1400,2386,596,532,802,6456,14388,5852,2042,875,1013,766,9243,1153,988,840,6619,1721,15031,662,2548,1412,1232,10781,555,4853,734,5937,514,4113,1901,765,789,731,3429,2546,8057,1745,3142,7667,1268,12485,1173,18108,768,10494,5599,2257,7533,4508,999,1607,459,1289,383,3468,538,675,4871,1591,12185,814,15092,2266,12008,3189,1599,3026,1642,1974,5516,1030,875,1356,2642,2936,2648,12265,1015,4309,9090,2528,12809,2353,2113,638,8501,1903,2092,1184,493,972,3448,2576,596,854,698,4956,393,1016,612,1224,663,854,1899,1820,2992,4653,1062,781,1359,723,2940,781,758,1687,4535,956,625,984,576,1953,1879,1813,675,738,7145,4939,871,1562,1669,914,2157,13445,1018,999,878,3249,2375,419,1060,646,4742,979,4269,760,2817,1193,418,668,758,10395,2291,14959,4429,919,698,1833,898,957,3089,2139,499,5423,1883,873,507,976,965,974,540,6793,16256,16297,1200,638,5610,5559,6333,3685,2398,941,2652,877,7375,683,7440,530,828,3589,969,2129,840,776,941,12146,787,1268,9209,1140,2051,884,713,505,779,1115,1591,6232,801,8818,1011,844,802,613,1949,2051,15116,1870,3336,18159,2857,678,943,1061,5889,667,5818,17366,6542,6104,933,9092,7617,670,459,1050,5063,7988,3304,741,1911,3005,2208,814,1359,4068,2030,1235,7633,5219,1815,1053,833,2338,1365,1076,2804,2357,1170,1259,1121,3653,828,511,1015,1655,701,2799,805,8972,8921,2331,766,607,622,1008,5908,1554,4513,10804,2371,642,886,1637,4623,776,802,13162,3192,4811,1442,1367,7070,752,3284,482,906,720,6578,4872,4602,838,1132,6352,680,1002,1003,5894,1956,5007,763,2715,583,544,1694,4167,8951,17659,1824,790,3011,5793,3012,862,842,14092,1758,8371,645,1744,432,2431,1402,2041,4973,1045,529,1760,1554,1229,2976,1318,734,6426,1358,7806,6201,10351,3101,707,4196,911,6261,1158,5250,1248,1419,872,1816,3697,4749,12082,1803,1427,984,16709,2846,1383,732,1573,672,796,1577,6403,3431,10345,703,3321,1034,791,4924,552,1187,6867,8922,1193,596,921,805,2357,5330,670,879,709,3751,2178,4038,13787,10082,1984,413,4877,2041,723,573,557,1095,7222,5062,905,4392,11086,1899,1857,8138,971,2321,737,5141,2475,872,931,684,7758,12883,7487,992,15974,3585,533,384,2839,732,1356,4435,1786,635,13109,964,12455,530,6852,5412,7155,16240,13178,7016,1682,3800,7577,15144,503,1817,1737,1438,587,13629,1569,1153,5352,2359,598,364,5215,3345,13949,706,4578,3084,5256,571,1187,795,3296,3123,18607,3105,4229,3135,667,9691,1668,3801,1949,5124,494,5105,864,593,975,648,1607,583,808,3987,1580,828,2752,10314,1259,2158,1644,2227,855,648,3274,645,2563,3710,1024,1561,984,828,972,1074,2801,15992,1408,10396,8485,1095,648,6806,2578,2898,8709,1142,827,2298,6588,1059,706,890,2645,4194,499,17916,1343,978,4516,5018,599,1665,814,5775,6676,1008,471,5046,2033,1170,975,4875,1211,1154,1767,16945,13250,2870,8848,732,1960,1622,15857,2924,758,1963,2224,4007,526,3942,951,1174,764,1001,487,517,5393,802,18578,4686,2330,811,17761,8069,3968,4827,1584,7515,4771,5143,3478,3758,1749,999,651,814,5960,1778,2891,891,2550,1875,1935,972,8286,3236,917,9972,1752,6391,8541,491,2177,2949,561,984,3097,2137,3505,10197,1656,2856,846,1192,1939,2233,540,1087,1151,1063,794,2010,6982,1689,1170,1689,1616,8400,10964,11376,628,545,1056,7943,5127,1896,596,1981,3089,2507,2496,810,917,1176,5882,15717,666,9214,12821,1013,1702,731,3464,3723,1154,825,11526,854,2143,1214,984,8093,12071,13653,684,8932,6849,2950,2052,1690,1243,984,1848,906,7513,1745,16558,3011,1720,616,517,5532,496,15852,10436,10406,1993,596,2162,1758,4764,1982,11051,2313,5412,1163,654,3454,7357,4228,1292,3183,552,5242,7893,1897,10646,2430,1865,568,5171,2420,477,1924,1240,4832,10546,730,5455,1698,10607,9408,713,1666,475,942,1956,1272,1115,3411,657,2025,727,765,1008,669,438,1949,1752,1240,865,4281,438,2949,1712,2874,708,2453,856,844,494,3530,805,427,583,1329,10454,5655,1564,456,4484,905,2303,1268,3030,6108,16337,872,3173,2328,697,764,782,2410,2347,3937,681,10228,790,2442,6239,9452,2793,7467,863,683,12720,741,5610,3090,5872,2575,743,1919,1041,7199,636,4619,696,1114,844,764,7059,896,1007,561,4971,422,372,5359,1659,766,854,13646,6904,1435,8639,684,16650,15035,944,1614,1435,7443,663,3759,5706,2258,4830,2085,2233,3195,905,1694,18648,2442,7805,872,2800,15246,1107,1410,12734,2839,974,876,6989,710,1343,975,1008,459,781,711,1812,620,805,1153,830,1450,10739,705,1244,6344,863,808,7999,489,673,1656,4355,15189,669,8896,5294,2255,449,686,441,942,1046,10038,5364,491,2047,670,3135,894,705,813,1055,993,2988,2347,16062,789,673,12171,1562,1792,362,4788,1370,1837,8334,967,10662,641,1864,2761,2011,816,2452,855,1846,658,702,932,1892,702,6453,6013,6762,1124,827,813,2594,1580,470,432,12338,670,1993,973,700,4427,815,3856,3475,803,872,6019,810,702,5653,1015,1956,7334,10636,918,895,6473,1024,2269,401,1223,1686,618,651,1327,3499,863,1132,2206,1814,5606,7451,716,1741,7978,802,464,1221,548,391,5646,2587,648,1207,761,14674,5269,3967,854,9471,1752,10283,12738,5967,1665,873,4795,658,956,1136,851,3064,1983,1103,789,10462,965,3528,1322,4243,827,5932,8341,6960,4032,1871,4466,5288,2016,4129,1169,1326,1085,14294,3192,1800,1715,1586,890,2960,852,1624,14968,3026,5698,1607,18440,15714,1610,900,1056,755,4478,1020,1700,2977,1664,449,1240,1111,7548,523,997,571,802,470,1065,13182,1744,1985,830,1401,1781,680,6088,852,864,548,13254,802,857,1384,868,1399,936,3671,1250,576,5938,956,480,776,1963,603,3864,1830,10947,776,17590,583,464,1161,3053,3290,968,3544,1721,13661,828,582,665,1410,8975,1043,816,958,1698,1235,1153,2578,3445,15218,15688,853,728,761,5125,853,5021,1177,2234,972,614,837,2564,4315,2466,851,13325,12648,5125,6467,1822,5002,593,645,4109,459,8121,4729,972,900,665,652,9533,1196,1381,3816,9218,473,7273,740,7128,2511,407,7290,596,1238,1607,11192,449,2442,4926,9203,3849,4183,1902,852,1033,1026,2642,1644,1003,11765,1893,579,2489,1228,1569,6160,1017,3553,17455,17569,6966,526,1443,11233,3455,15851,1715,963,1662,954,3176,2583,3190,1048,10935,10937,4596,3667,787,5291,1031,1289,7372,3278,587,521,1207,2406,988,1662,5067,725,1031,4639,2211,1356,16599,777,5697,2368,4981,5455,7966,3071,984,4939,8930,1595,2096,5167,10313,719,4319,1838,761,1624,1961,1893,6573,495,868,1289,972,7325,2184,6036,12554,7400,805,454,644,2104,1412,7942,654,622,14027,721,779,1059,2644,577,432,1991,2883,2710,6199,3701,1097,1882,3796,11584,695,434,932,5252,9581,2159,804,844,16261,2692,863,3468,18395,1031,18756,6112,1427,758,421,975,9734,1949,14593,3360,532,12219,10907,3049,1963,10886,2647,844,8698,463,2386,8752,828,1881,1626,1020,764,3162,3655,3435,6039,4486,1591,4033,766,772,571,970,2426,802,891,752,491,2744,1048,2717,820,4864,969,2841,3913,2329,1115,3478,8213,1166,1624,11147,1307,6751,3871,788,775,1208,2699,1657,2117,8674,702,841,5042,2359,893,1279,2394,1401,5006,17889,5220,1343,6968,734,631,4044,4704,603,17688,830,945,956,942,2271,789,4883,1975,492,6648,2064,14386,942,561,1809,17029,4584,918,2157,1370,1026,1436,2319,799,2496,6682,4412,518,3763,2217,8233,949,1103,526,469,1827,2923,2387,8227,1454,8774,945,990,1172,2822,4186,2699,3810,6857,708,7167,12677,1878,755,3381,4854,2609,1004,5805,8982,6288,4180,4233,5319,14542,789,761,776,5296,1807,2150,2789,3176,918,2795,1092,902,3197,743,18055,827,2079,669,838,507,713,745,3220,1215,2407,15247,438,926,15231,863,1118,861,1165,5044,1035,2591,763,784,734,745,9013,7752,6480,5189,491,539,6797,872,652,1274,1052,2324,6737,1815,418,11119,545,5792,1394,617,4290,3694,1235,3103,4030,687,2658,645,5363,1436,540,2596,5163,506,5443,1786,7247,461,842,1778,470,2131,942,912,13994,936,15197,1832,2551,1046,1002,3478,9796,1114,5439,5897,1662,720,5312,18293,5841,1299,1624,863,11525,904,864,926,2016,678,524,4101,2680,523,3603,7870,2414,687,1857,9533,734,9641,422,2972,1649,17882,589,1334,838,15968,1743,1588,7655,2078,1057,1131,898,2719,3299,5038,15511,4771,4653,7466,13512,608,12958,5743,3226,1229,1089,754,2079,4558,1059,4668,906,4001,1431,1682,1947,448,5308,898,15324,767,915,738,6295,9289,756,1754,900,867,1600,2306,781,3557,4946,3802,882,5233,2919,1030,1723,1923,13872,5458,1257,4749,1822,12047,776,1000,1050,1257,3256,2184,8529,4530,1052,1662,663,1873,785,948,4789,3502,745,2610,2365,462,10742,3035,2607,1308,847,1353,2834,14138,878,1671,7589,1621,12725,15272,1014,1018,1334,663,590,4537,984,1177,848,605,1668,4196,2556,1644,7600,921,868,1046,2016,1694,945,8275,2529,18440,5884,2209,3601,2527,3342,877,684,3183,484,435,931,1133,702,743,690,833,4162,873,579,4791,3941,1173,743,459,15843,706,886,9461,10389,800,17534,1333,2877,531,2442,9572,2901,5599,6303,2815,2781,660,936,5804,1205,3012,470,1004,2801,2557,1656,3284,3562,950,4439,9245,4779,521,1243,6196,1715,5707,5838,3075,6458,6980,1750,1628,480,515,1817,6062,1060,1421,2295,2819,4013,6411,9694,2018,984,4627,2958,1691,2269,544,949,2724,1882,1847,778,777,1848,1018,1168,5031,943,3922,8303,2082,10738,984,6509,477,1254,1250,8119,1810,4223,5639,1881,2386,949,544,505,828,3121,936,2230,612,4666,3035,15955,684,952,1266,419,625,1935,6928,481,1445,3791,5154,946,3247,10051,658,11434,1797,3781,2237,4241,4679,552,6857,1916,515,2313,886,1122,689,1264,9281,8596,707,1781,992,2927,4323,1030,12947,10309,4560,645,13406,925,534,1698,5241,13912,7238,1624,707,730,9439,633,1915,928,798,1245,2624,1355,723,5969,692,545,411,7480,911,538,10577,433,921,3618,1051,6887,15303,702,3462,1782,1888,2301,16538,457,707,915,1079,1046,4504,14482,839,622,624,6377,4724,923,5816,9230,7014,819,18363,766,1612,931,626,983,579,524,794,3332,18172,828,1238,1013,12848,5670,949,11530,1884,7081,16492,787,4127,870,10093,610,3590,1809,15134,1680,8014,5107,1076,1085,13588,12320,17747,886,688],"type":"box","orientation":"h","name":"Ideal","marker":{"color":"rgba(253,231,37,1)","line":{"color":"rgba(253,231,37,1)"}},"line":{"color":"rgba(253,231,37,1)"},"xaxis":"x","yaxis":"y","frame":null},{"fillcolor":"rgba(93,200,99,0.5)","x":[2467,12343,4525,4326,7274,683,5824,4200,5219,9882,1103,4579,16137,5817,16733,6089,5761,11200,6969,1395,4974,523,1863,7056,1000,1944,15110,5958,2686,2839,2827,14199,5292,2172,5622,2381,702,5281,9256,9271,14196,1116,4872,1559,4363,2616,1749,4256,526,3355,2391,3787,3819,4969,2670,8408,2686,7277,5450,4689,1436,3512,523,11975,5821,11021,1792,7560,5468,926,6652,11360,13786,1116,3676,2590,906,3312,1438,4381,3402,1378,6885,4309,4238,919,1225,4657,8145,5893,2135,906,3674,9157,1627,938,3519,702,1282,540,4173,1052,684,710,941,907,949,854,6383,4989,4492,2516,1206,11322,7182,3082,2020,6405,5292,7741,1069,7695,15841,645,393,5331,4189,13853,905,2536,5967,1035,3266,1743,3250,1002,1125,936,921,867,1928,579,4949,6532,1063,2801,1024,596,1122,11255,5226,734,6075,4811,1909,628,4064,623,13540,686,5242,4514,4212,2636,13387,5269,2874,2879,18026,5845,4124,1196,14483,4365,10688,6047,5204,2486,854,9659,3167,1061,3555,8879,5376,698,12338,803,10706,1076,716,12308,9836,16409,5581,2314,10043,12418,574,3713,2112,7781,4732,608,1316,9253,4751,698,3437,5869,1103,4838,2847,15662,3347,1273,537,11177,4670,957,5102,4815,4357,2348,2935,1067,7553,4168,4106,1263,2348,4946,394,9478,2802,1708,936,731,18706,6887,743,734,2826,13092,4538,743,911,8816,2735,14465,2012,878,1010,5410,10308,9205,8020,1433,730,7123,6244,9728,646,960,5288,6115,8688,965,4770,830,1026,828,3293,447,1125,4873,17891,802,4900,4096,645,11825,945,3704,5729,833,16336,5561,9793,4953,2753,1917,1064,6206,576,3790,6037,1402,10584,1395,1845,5624,3832,1238,1819,3998,6316,7589,15568,1583,10388,6278,9882,4513,14982,6026,3205,625,906,9905,2588,1094,10800,878,6547,7632,5996,737,4044,5312,957,844,5602,4379,1012,650,775,1142,1080,1096,666,4559,8721,593,1186,5103,15109,2264,12081,7580,3562,982,3472,990,673,5241,4308,900,1556,1314,5291,7392,1125,11654,1624,1645,1428,2271,10211,1115,5925,2373,4578,1746,9596,802,828,4936,3603,526,8829,994,3311,3296,6056,10389,743,1046,828,4718,765,3291,2567,16198,579,2239,1882,5268,628,2790,4482,524,803,17334,7287,6564,4914,1656,7414,4620,18795,2828,802,518,2316,521,1015,4746,1011,5468,7275,726,3180,1035,4332,11666,844,3214,765,404,1607,1743,5975,4642,815,6499,7486,5002,15562,666,765,827,5557,4793,10302,855,8686,1038,14247,5044,1003,7849,3924,877,473,5523,561,957,1813,5939,2863,4788,16390,810,3002,918,605,879,3604,1940,2089,15797,576,969,4144,12862,1117,2761,2777,12196,13543,6319,941,1656,3377,2043,2352,6756,8654,16643,616,5164,3740,945,6899,5088,1081,4497,7899,8624,4164,743,1662,2708,5430,2200,810,593,1041,880,1654,930,9137,4037,773,9025,1619,6097,7179,8888,14611,3914,1046,16854,806,7086,1082,3669,14179,10580,3648,3380,1420,803,4478,1056,1085,596,988,3921,11188,4819,9738,17513,4939,855,612,9541,2699,828,1766,7246,1238,12549,658,2629,18255,576,596,1053,5550,710,3502,417,13420,646,14543,12707,574,1323,15188,576,4446,6618,2484,15654,3024,4355,7162,4791,5556,5698,863,803,6629,3095,10053,2190,4062,16778,3271,827,17209,8501,13293,5058,746,1024,668,16872,4137,787,4562,13027,7812,1760,1381,5980,4480,1624,10738,1571,2479,2393,5799,9710,1882,2415,816,408,4601,17579,730,746,1080,12644,608,3427,612,942,993,2295,5487,9574,675,727,862,7006,2484,7963,756,2616,2002,734,2981,9537,5058,648,17073,506,4718,791,5174,847,5946,3387,698,1206,990,5151,3405,10888,827,7553,11796,17849,2711,900,5247,7534,4368,928,3096,729,8836,6814,3941,5437,7603,1211,1610,3761,9555,6406,2446,435,2805,7003,864,4973,14833,13248,5973,473,675,593,648,5264,3506,405,9768,2939,568,7518,7466,3959,3925,4315,2328,5171,4586,1148,11106,2285,1910,797,608,788,1953,4899,3745,658,624,8743,4480,4472,698,2762,1082,2387,945,4688,2863,1362,4612,6066,2275,2040,4879,6743,12968,720,666,2215,2354,9704,15105,5218,3385,11871,3043,5140,2184,4844,1656,876,2821,1303,4476,454,4989,4830,878,1000,15451,1279,5569,17522,2562,776,4890,4088,3006,13720,3175,689,8476,743,2369,516,17634,935,16169,6296,4772,18426,5898,8128,1439,4702,5167,2115,1914,1154,3013,3740,11544,3150,775,6335,16626,770,10012,12831,7305,10539,844,2148,666,942,2999,1389,2897,4471,12499,5408,947,8127,2913,982,18741,3455,537,743,2196,17825,1740,1063,2489,3352,693,10401,5100,1167,9081,2175,8183,1842,900,772,1801,1026,5699,2428,5142,5889,1363,11549,675,1009,4965,951,5940,8540,12677,1637,741,7220,4562,1989,2064,12071,998,748,7677,1059,1007,4740,12832,7931,842,2777,16779,963,11644,7079,4742,2239,6425,1050,1140,9513,1139,612,8930,6108,1687,1966,557,2425,507,518,983,10424,2913,7466,579,1115,7032,6479,544,6751,4229,3215,984,1738,767,1728,9408,10090,3749,3808,5717,1875,574,7550,4611,530,2112,1088,12747,5912,1073,6588,4318,1689,936,731,1921,6776,1114,706,625,755,5433,9720,579,1316,11088,13098,3266,2801,16309,4284,898,5292,2331,7875,734,645,7955,4242,6416,4158,1153,5134,4584,4858,635,816,800,689,1179,8820,1827,1599,5504,488,1219,1605,3496,1046,17235,9122,8366,868,4816,5557,730,2371,12338,17216,3353,828,2657,9961,593,1845,666,668,3920,614,720,2939,789,1179,2176,639,8316,16314,6140,1949,1914,5301,756,906,2148,2760,6091,9697,5880,5606,1232,5739,17849,8114,2399,9377,4284,4230,9419,17294,18419,2753,765,3078,540,7408,17650,4222,17153,645,1274,834,628,553,7811,10274,1815,12210,7844,421,1600,6989,700,7258,4200,1613,774,789,5718,550,2865,8110,2037,6499,4578,3774,11078,16957,933,17987,1002,1814,1698,2717,10115,6588,14201,3018,7610,12182,7611,687,2814,746,984,2443,8113,1551,11279,6313,7056,394,7918,3917,12236,4562,1050,965,4774,1606,7679,2392,4155,10512,2329,2549,5518,4916,2711,6188,2467,394,14024,995,16277,10091,5140,4468,3198,4743,3801,1935,15065,608,2815,13052,2469,689,17203,14386,13753,7351,6470,5421,2352,872,4183,3965,1142,2647,1002,1026,1041,851,8808,988,842,507,1613,1040,2137,2153,793,4096,15984,2469,2826,802,765,2838,574,5592,1270,540,13037,13813,773,3618,8281,737,1129,4719,2035,647,1676,4435,600,786,2646,9877,984,2657,11382,3133,3996,3370,561,11105,3360,9525,3006,743,5556,10388,3091,2447,4682,4129,15095,833,5164,4374,2451,2329,14803,1024,990,602,7848,7577,1838,1662,1071,6516,10337,4751,1727,4077,1356,4688,2579,790,3775,12787,1433,713,5042,8067,780,506,14488,2360,596,5267,4980,9952,1577,700,990,708,8521,805,5285,907,716,612,1192,7715,1819,1957,4687,1332,4838,2718,5012,8964,891,470,6156,1415,11301,1799,649,6694,3854,6012,574,4268,1588,5169,2287,743,1415,1061,4090,12437,2300,6442,17175,3546,1358,842,6337,3105,3545,1013,3656,6377,5087,817,603,9541,13034,1046,10428,8314,7455],"type":"box","orientation":"h","name":"Premium","marker":{"color":"rgba(93,200,99,1)","line":{"color":"rgba(93,200,99,1)"}},"line":{"color":"rgba(93,200,99,1)"},"xaxis":"x","yaxis":"y","frame":null},{"fillcolor":"rgba(33,144,140,0.5)","x":[1668,4640,5755,17422,685,2100,12829,4002,2643,1374,8312,3950,682,3748,5783,12845,6975,596,1143,734,7747,1569,16670,953,3472,645,5868,16116,460,4508,4081,2501,3160,453,5938,8637,2367,710,8073,4084,3061,1187,478,1771,10962,7145,1662,2125,692,402,2656,3664,628,7350,558,648,10147,3114,4031,647,3724,4760,1031,4544,13015,579,2130,554,470,6539,419,6204,368,4594,17153,552,6563,429,619,6786,2282,4306,2838,548,828,593,784,4826,8475,3278,657,3307,18118,526,5919,14561,622,821,7438,2442,7333,1814,12451,1799,4972,17247,765,3774,14527,480,3465,489,462,855,3231,9257,554,608,1869,644,978,4498,4068,6221,610,3145,10165,11390,13782,3015,593,675,8214,5070,3734,6167,13068,2145,2753,2106,4286,3590,2222,6075,18257,1686,756,656,15092,8815,2040,584,14740,6733,530,2369,3184,1273,15618,14433,7677,478,1372,898,3833,1128,2673,2811,15769,2932,421,449,3360,847,1062,2264,4583,1078,1945,7257,926,1883,492,967,435,2573,2198,5671,5522,588,2093,5265,2784,5372,6563,2368,1552,7125,14931,3045,7487,13215,4064,433,700,3368,421,407,9642,5160,540,4379,2956,8794,811,1030,6079,5638,3700,1907,3601,6245,6486,2453,1785,2862,10285,548,3465,6654,1091,4063,871,1238,504,3752,2161,3107,2482,1154,5190,645,1706,4045,5050,2512,6814,1777,1378,1626,4580,691,9094,487,10546,4281,432,2893,4279,724,733,15773,642,642,1717,3178,2036,4657,3276,14386,1367,954,675,421,3989,12175,886,3549,737,7567,1607,1436,4693,694,1833,4973,3407,4570,2426,505,5750,518,2590,2326,2564,2819,489,1193,2048,819,7583,4068,571,17694,453,730,521,6892,666,958,7747,14527,3298,868,3789,6566,2655,645,3614,13940,597,2338,687,14768,7891,1154,6168,4959,1894,774,9609,4214,5919,10113,540,2726,4584,566,7001,7372,462,1648,4292,4797,745,2797,7703,686,2753,1074,2473,15334,5025,702,1171,4187,1007,596,524,17920,8895,1600,4559,16677,675,5236,3340,1799,1066,5174,2878,4298,707,13080,1316,400,1105,3340,408,631,932,434,3177,3181,1939,1867,1111,5245,6518,505,12000,3795,2553,897,1709,8662,4588,633,2949,4766,1805,2694,4414,530,5010,2508,2618,18127,624,478,2537,2753,4412,714,1348,4659,1552,12714,3996,5324,12916,878,945,14217,17871,3518,4738,674,2854,1744,933,2280,1008,575,2329,2889,4309,2936,4129,2484,3473,382,2008,6459,7910,6726,3398,3324,12381,537,645,720,5293,2764,7343,646,3296,2826,1340,3888,4297,6105,462,3593,756,3464,3180,652,7507,4276,681,9043,631,421,1741,2138,552,436,773,2423,5152,5565,545,2745,802,814,625,8983,9523,9009,625,4749,2259,7841,4473,3783,3295,394,2102,1304,1365,6301,4520,447,5381,421,679,1641,1748,6962,1657,2880,1986,3167,3323,552,4432,2551,13564,945,789,3738,10851,3378,507,1154,3966,8184,4210,6103,880,7903,1021,613,783,683,526,2569,1140,4576,2404,530,2219,10134,6996,12196,6169,2656,608,6019,2807,4282,794,821,664,6132,3895,544,5953,1853,722,15510,1419,620,3394,18067,842,4282,789,4162,7094,2312,2776,13669,2444,462,1079,2106,6534,9474,3437,9089,7774,3755,2564,2042,612,3535,2674,4933,526,2481,880,6094,450,2309,608,13500,2382,705,939,608,1814,12459,13499,5600,5799,625,5688,499,4762,4688,694,9556,1569,11189,7173,757,2945,4181,7010,701,2585,9576,921,837,3345,4341,790,6663,4662,4200,642,1621,3124,706,13043,2112,4690,3249,13111,1244,6288,14066,4466,1623,16111,717,9158,1836,487,15466,11688,7676,720,645,1387,5533,4879,1190,683,8261,2437,906,5174,6701,511,2505,1083,680,5351,18176,7673,620,6239,523,765,4851,579,1024,773,2681,478,2617,7139,2647,362,17724,2095,12907,811,1565,3888,4418,432,10204,6073,2753,521,7136,3673,2889,3293,2228,548,645,2832,1151,3160,921,5017,4665,1167,10032,1116,12581,9705,4108,3407,9904,3872,11382,1351,1628,3350,12141,3226,5950,2686,4195,485,3570,5016,485,2923,7415,492,12430,1624,4633,6335,9513,6702,1132,1160,2207,3685,2973,2475,434,8994,4724,4514,9960,3264,664,1200,7706,4755,697,3806,2200,3447,759,8505,1438,751,4232,1187,2808,847,771,1615,793,419,9510,11214,6031,4869,13161,1187,651,4821,552,4447,432,895,7730,2356,4311,477,658,668,2428,11146,2467,1401,737,1264,850,526,1442,5593,3418,1271,1436,4395,2482,4315,679,4032,614,1637,1107,1757,5505,1875,5229,1914,487,1727,4191,9058,485,3606,8931,5914,336,2400,658,5941,391,1079,944,6296,14603,4754,597,4253,4816,4435,17425,447,952,2227,18429,1717,5002,1569,8032,5241,1392,561,1373,2120,1194,756,7453,1180,4712,3924,801,2319,18398,1821,393,6777,789,5480,6871,1075,6539,7797,1240,3118,8257,6195,2382,4697,2688,900,3010,7076,5346,530,2866,1918,11392,17955,1777,4039,2095,11764,17084,16353,13631,5119,10771,16582,4565,569,2314,914,521,5183,7662,898,11946,4295,4077,1239,530,2261,1569,737,6781,505,4252,1367,2570,5065,1237,17279,2426,589,1198,5422,2445,1237,10993,6500,6140,11586,2601,7131,730,552,384,4507,10824,1592,2765,3799,2933,5067,373,3061,904,4588,8627,4173,679,1031,795,1393,473,6285,633,3462,4191,7942,1655,3462,561,4466,852,1098,3160,850,6669,3599,2400,2841,6438,600,2066,8167,814,2345,810,2088,3890,1745,754,449,1550,4563,1053,1666,1408,5759,1179,2104,5672,8151,4189,463,4564,12621,1236,521,1291,5759,11957,2623,7637,10023,6662,1867,505,2705,4385,9278,6825,7589,9552,4383,505,3037,5819,4078,726,3063,3991,1227,1689,633,2039,2372,4853,4092,4498,3624,2036,16041,8138,12981,945,2441,1247,2218,921,889,1184,2517,682,868,10915,14792,10226,4497,702,710,878,3567,7258,1958,5038,2005,551,1676,868,1829,3057,620,16874,1875,758,8349,755,4619,766,477,12862,14236,5387,3731,2095,12874,2400,2481,1172,9797,938,633,684,729,767,4503,1281,645,5484,5557,9680,7127,1129,3605,573,743,12267,4124,4123,4921,6132,4514,9896,3763,427,6125,12799,9140,711,4082,5645,3350,2401,1410,1115,9780,13099,4321,2009,7677,1424,1632,4484,2345,5450,957,912],"type":"box","orientation":"h","name":"Very Good","marker":{"color":"rgba(33,144,140,1)","line":{"color":"rgba(33,144,140,1)"}},"line":{"color":"rgba(33,144,140,1)"},"xaxis":"x","yaxis":"y","frame":null},{"fillcolor":"rgba(59,82,139,0.5)","x":[3250,1771,6097,698,4639,9173,472,965,6250,4296,4844,2926,607,3819,776,4816,2751,1643,5056,3959,13853,1775,783,4025,8973,3634,1354,645,12117,1935,1841,4427,3253,10428,4543,7114,15649,2700,2871,4416,8868,4528,4480,2229,1821,4766,625,1172,7306,8430,755,2534,2516,4265,548,5443,669,7368,3906,4899,3050,11512,3839,10886,3734,13603,1348,13068,2604,796,7190,3163,734,2898,3672,1662,6429,1202,6738,2307,11577,5226,705,7049,1897,2026,540,5420,8313,5505,5818,1410,3360,733,3063,6770,3363,3775,492,641,2074,2290,2708,8731,6666,1758,4569,3965,591,4737,589,2320,8104,3072,1213,2150,4060,6851,3752,8732,4506,650,9741,3399,4389,6443,4824,558,3895,1190,4475,4620,2247,3669,2642,7665,6335,1146,1332,394,12121,12467,2753,684,791,739,1147,5458,3852,4389,2906,3348,4312,11854,5443,5786,612,13978,895,335,3750,4497,6750,3640,421,1138,8107,1090,5600,16690,12754,2854,1951,10164,522,11576,15977,2498,492,456,3436,3744,1042,1343,515,7026,4171,1433,1443,548,738,707,844,4189,473,414,1333,8048,17869,3307,2327,1268,5979,681,816,1800,4445,9380,4672,700,6686,2234,435,1254,836,13320,1443,1594,2017,8569,1289,1407,830,2161,645,6517,600,1968,2599,610,3485,5701,2288,4131,5497,4026,827,2810,571,505,14027,6793,7207,3923,2960,1190,1821,653,1625,6963,6271,1637,789,4749,2331,3484,5526,537,4517,3948,3946,7143,2680,450,3570,2394,3024,596,2058,8280,4006,7251,4257,652,4221,684,4596,465,3660,1752,969,457,1760,6407,4381,1172,16737,749,4851,3878,9281,8736,584,5492,4937,3105,1121,4320,394,4566,6042,4095,5242,4633,8013,3595,620,4004,801,12121,3626,2258,6743,628,675,5893,6925,4150,3620,4620,5132,2863,11954,2100,9609,653,9569,4504,5863,1111,4582,6662,492,5817,2873,506,780,490,2827,6798,11235,5010,1128,4807,2212,4440,1286,777,2642,3453,4847,1257,2270,1333,6330,7260,4508,5219,3107,612,3663,6020,2511,2239,489,5825,1175,2066,5005,432,912,14745,6966,2074,5378,4155,10184,394,3979,4844,3787,1872,4679,5198,596,477,593,1637,2130,504,8492,2583,7162,15467,2278,470,447,11554,7308,526,6982,5002,3884,3811,3304,2676,3042,487,5544,3884,4712,473,4538,1262,776,11096,5330,4081,7929,669,532,544,9526,8220,3730,564,2795,4149,4203,2877,13903,1374,949,517,2344,2428,2517,3595,3312,925,3506,3889,684,4140,4826,485,2751,5412,2494,3376,2298,1715,3210,4353,4398,581,18236,2015,816,678,3489,2427,462],"type":"box","orientation":"h","name":"Good","marker":{"color":"rgba(59,82,139,1)","line":{"color":"rgba(59,82,139,1)"}},"line":{"color":"rgba(59,82,139,1)"},"xaxis":"x","yaxis":"y","frame":null},{"fillcolor":"rgba(68,1,84,0.5)","x":[8818,1881,2376,1323,3282,2500,13853,3869,1811,2788,2596,3734,3382,3002,2732,9095,2686,5622,4630,675,1865,727,2290,3862,945,3613,2037,4403,18565,2381,4118,4129,8716,1092,2239,1103,1791,6340,1806,1098,5919,6455,6250,2375,4525,1890,3039,6108,4887,3304,4423,994,4536,3920,4038,1737,3062,1027,6838,4751,673,2100,3296,6727,6397,2167,593,1949,4309,4861,2518,8260,2310,3104,1001,10562,1974,1273,4844,5430,2396,1564,1577,4725,3136,2045,827,4328,1997,5334,1389,7333,12923,3080,3802,3280,15824,3136,875,4047,7476,3172,3669,2912,1166,8996,2912,7098,1749,2301,3077,4939,1235,10577,2443,10217,4191,6564,12610,662,722,1155,2805,3437,4554,3871,5171,4113,13502,1323,1808,4277,3265,2293,3965,563,1808,4140,8216],"type":"box","orientation":"h","name":"Fair","marker":{"color":"rgba(68,1,84,1)","line":{"color":"rgba(68,1,84,1)"}},"line":{"color":"rgba(68,1,84,1)"},"xaxis":"x","yaxis":"y","frame":null}],"highlight":{"on":"plotly_click","persistent":false,"dynamic":false,"selectize":false,"opacityDim":0.2,"selected":{"opacity":1},"debounce":0},"shinyEvents":["plotly_hover","plotly_click","plotly_selected","plotly_relayout","plotly_brushed","plotly_brushing","plotly_clickannotation","plotly_doubleclick","plotly_deselect","plotly_afterplot","plotly_sunburstclick"],"base_url":"https://plot.ly"},"evals":[],"jsHooks":[]}</script> --- # **plotly** package (2) Another example with a scatterplot: ```r plot_ly(diam_ech, x = ~carat, y = ~price, type = "scatter", mode = "markers", hoverinfo = 'text', text = ~paste('Carat: ', carat, '\n Price: ', price, '\n Clarity: ', diam_ech$clarity), color = ~carat) ``` <div class="plotly html-widget html-fill-item-overflow-hidden html-fill-item" id="htmlwidget-db4c99f7f7192c34b7e0" style="width:360px;height:288px;"></div> <script type="application/json" data-for="htmlwidget-db4c99f7f7192c34b7e0">{"x":{"visdat":{"13c6a22d0b9922":["function () ","plotlyVisDat"]},"cur_data":"13c6a22d0b9922","attrs":{"13c6a22d0b9922":{"x":{},"y":{},"mode":"markers","hoverinfo":"text","text":{},"color":{},"alpha_stroke":1,"sizes":[10,100],"spans":[1,20],"type":"scatter"}},"layout":{"margin":{"b":40,"l":60,"t":25,"r":10},"xaxis":{"domain":[0,1],"automargin":true,"title":"carat"},"yaxis":{"domain":[0,1],"automargin":true,"title":"price"},"hovermode":"closest","showlegend":false,"legend":{"yanchor":"top","y":0.5}},"source":"A","config":{"modeBarButtonsToAdd":["hoverclosest","hovercompare"],"showSendToCloud":false},"data":[{"x":[0.73,0.7,0.31,0.31,0.31,0.83,0.51,0.7,0.4,1.1,0.52,0.32,1,1.01,2.01,1.6,1.01,0.31,0.38,1.03,1.31,0.7,2.01,0.8,1.2,0.31,0.82,0.41,0.33,0.33,1.3,0.35,1.25,0.62,0.3,0.4,0.9,0.4,0.5,1.09,0.91,0.34,1.66,0.42,1.17,1.27,0.9,1.55,1.5,1.06,0.9,0.42,0.32,0.38,0.78,0.3,2.01,0.34,1.31,0.9,0.33,0.31,1.22,2.15,1.27,1.06,2,2.01,0.23,1.16,1.01,0.34,0.55,0.43,0.5,0.3,1.52,1.22,0.3,0.5,1.26,0.41,0.31,0.5,0.39,0.31,1.3,1.05,1.06,0.51,0.52,1.52,0.31,1.33,0.41,0.4,0.88,0.32,0.7,0.53,0.52,0.69,0.65,1.2,1,1.2,1.5,0.38,1.51,0.35,0.3,1.01,0.54,0.31,0.5,1.04,1.19,2.13,1.1,0.3,1,0.71,1.02,0.51,0.71,0.55,0.34,0.79,0.36,0.3,0.89,0.57,1.22,1.09,0.54,1.02,0.9,1.14,0.26,0.54,0.9,1,2.02,1.5,0.53,1,2.3,1,1.51,0.62,0.81,0.33,2,0.31,1.01,0.57,0.77,1.03,1.23,1.24,0.71,0.31,1.04,1.19,0.41,0.4,1,0.53,0.27,1.11,1.63,0.9,0.42,1.73,0.41,1.3,0.24,0.35,0.3,1.01,0.27,1.16,0.59,0.7,0.5,1.5,0.55,0.36,1.05,0.3,0.9,1,0.51,0.81,0.72,0.33,0.23,0.51,0.84,0.59,0.91,0.52,0.87,0.3,0.96,0.63,1.09,0.92,1.01,0.3,0.31,1,2.2,1.01,0.71,0.72,1.31,0.77,0.51,0.59,1.54,0.57,0.71,0.51,1.31,1.01,1,1.5,1.06,0.32,0.34,0.31,1.11,0.41,0.72,0.32,0.53,0.74,0.4,0.7,1.5,1.13,0.5,0.41,1.04,0.92,1.5,0.9,0.31,1.14,0.58,1.57,0.91,0.7,0.7,0.92,0.3,2,1.32,0.58,1.05,0.7,0.35,0.7,1.5,0.58,1.01,0.31,0.32,0.7,1.52,0.7,0.7,0.92,0.9,0.34,0.33,1.5,0.42,1.05,0.31,0.7,0.32,1,1.02,0.27,1.08,0.43,1.13,1.5,0.3,0.56,1.2,0.36,0.33,0.51,2.07,1.01,0.33,0.6,0.6,0.35,0.75,1.12,0.26,1.01,0.43,0.27,1,1.03,1.01,0.33,0.24,1.01,0.3,1.02,1.5,0.5,0.3,0.71,1.11,0.28,1.29,0.57,0.33,0.75,0.35,0.32,0.32,0.29,1.03,1.7,0.9,0.32,0.61,0.51,1.13,1.02,0.3,0.31,1.51,0.81,0.7,0.74,0.51,0.35,0.62,0.32,0.58,1.01,1.07,0.7,1.04,0.51,0.53,0.3,0.9,0.78,0.33,1.51,1.02,0.43,1,0.38,0.6,0.54,0.49,0.9,1.23,1.21,0.7,0.31,0.56,1.71,0.25,0.35,0.32,0.4,0.82,0.9,0.41,0.3,1.5,0.76,0.51,0.4,0.7,0.24,0.31,0.31,0.73,0.41,0.28,1.17,1.5,1.5,1.26,1.18,1.02,0.26,0.4,1,0.35,1.01,0.34,0.73,1,0.45,0.71,2.12,0.3,0.4,0.32,0.31,0.33,1.01,1.52,1.77,1.08,0.33,0.32,0.27,0.32,1.24,0.36,0.71,0.9,1.47,0.56,1.8,2.02,0.3,0.38,0.31,0.91,0.39,0.49,0.31,0.55,0.33,0.91,0.73,0.42,1.3,1.24,0.72,1.01,1.72,0.72,1.21,0.33,0.71,0.7,0.73,1.5,1.01,2,1.5,0.71,0.34,1.29,1,0.72,0.3,1.17,0.5,0.7,0.3,0.31,2.04,1.33,0.32,1,1.52,0.71,0.3,0.33,0.34,0.34,1.01,1.52,2.02,1.64,0.4,1,0.38,0.3,1.1,1.08,1.02,0.84,1.51,2.01,0.71,0.32,0.32,1.19,1.01,0.71,0.31,0.41,0.7,1.25,0.87,1.5,0.59,1.01,0.36,0.31,0.78,0.7,0.39,0.7,1.07,0.4,1.04,0.36,0.81,0.52,0.31,1.1,0.41,1.07,0.73,0.23,0.32,0.26,0.3,0.33,0.4,1,0.73,0.71,0.38,0.39,0.41,0.35,0.55,1.01,0.42,0.31,0.43,1.12,0.51,0.52,0.3,0.44,1.01,0.9,1.56,1.01,0.33,0.3,0.9,1.7,2.02,2.17,0.28,0.53,1.59,0.5,1.07,0.53,0.42,0.33,1.08,0.32,0.44,0.73,0.84,0.41,0.32,0.31,0.3,1.51,0.5,1.04,0.34,0.31,0.41,1.51,0.91,1.22,1.2,0.51,1.51,0.81,0.31,1.24,1.01,0.36,0.7,1.04,0.5,1.02,0.31,0.41,1.12,0.42,0.27,0.68,2.14,0.53,0.7,0.31,0.7,0.34,0.41,0.57,1,0.32,0.74,0.9,0.53,1.3,1,0.5,0.95,2.01,0.51,1.11,0.75,1.5,0.72,0.74,0.7,1.01,0.24,1.01,0.43,0.43,0.41,1.24,2.03,2.01,1.23,0.78,0.4,0.63,2.32,1.12,0.56,1.59,1.04,1.01,0.32,0.7,0.42,1.5,0.32,1,2.04,1.05,1.53,1.48,1.12,1.01,0.82,1.51,0.56,0.3,0.25,0.93,1.08,0.6,1.53,1.5,0.4,0.9,1.03,1.23,0.84,0.71,0.26,0.37,0.52,2.11,1.51,1,0.55,0.51,1.01,0.32,0.58,1.26,0.78,0.84,0.23,1.5,0.33,0.51,0.73,0.52,2.04,1.54,1.53,0.24,0.55,0.52,0.44,1.19,0.35,1.76,1.01,0.41,0.76,0.5,1.09,1.21,0.52,0.39,0.5,0.92,0.37,0.7,1.71,1.21,0.52,0.51,1.2,0.31,0.41,0.81,0.72,0.83,0.63,1.8,0.32,0.34,2.14,0.7,0.3,1.21,0.54,0.41,0.43,0.32,0.24,1,0.51,0.8,1.54,0.74,1.23,0.32,0.29,1,0.41,0.41,0.55,0.57,0.39,0.4,0.82,1.5,0.7,0.34,1.03,1.03,1.04,0.61,0.31,0.72,0.98,0.29,0.27,1,0.38,1.83,0.51,1.16,0.5,1.52,0.91,1.04,1.02,1.01,0.7,0.31,0.4,0.65,0.7,0.3,1.02,0.32,0.71,0.33,0.38,0.51,0.3,0.54,1,0.3,0.31,1.51,1.02,0.31,0.76,1.12,0.71,0.55,1.12,0.58,0.41,1.02,0.71,1.05,0.33,0.7,1,0.3,1,1.5,0.86,0.26,0.35,0.5,1.5,1.01,0.32,1,0.39,0.41,1.2,1,1.01,0.73,0.31,1,0.33,0.58,0.93,1.31,1.04,0.42,1.04,1.02,0.79,0.38,1.01,1.01,0.33,0.67,0.44,1.23,0.52,0.7,0.8,0.33,1,0.37,0.74,1.2,0.31,0.71,0.51,0.76,1.27,1.7,0.31,1.53,0.83,0.52,1.06,0.53,1.51,1.51,0.71,0.34,1.52,0.9,1.54,0.56,1,1.01,0.32,1.2,0.75,0.62,1.16,0.82,0.3,1.52,0.91,0.37,0.32,1.11,1,1.01,1.5,0.58,1.06,1.5,0.32,0.91,0.51,0.31,0.38,0.27,0.9,0.78,0.55,0.44,0.7,0.54,1.04,1.04,0.7,0.43,1.04,0.32,0.8,0.38,0.5,1.51,0.5,1.19,0.94,0.45,0.32,0.4,0.25,0.79,1.52,1.22,0.43,0.31,1.08,0.43,0.7,1.21,0.3,1.22,1.21,1,0.74,1.5,1.64,0.93,0.31,0.91,0.37,1.21,0.73,0.3,2.02,1.21,0.4,1.06,0.3,1.57,0.72,0.39,0.41,0.38,0.31,0.65,1.32,0.9,0.32,1.2,0.46,0.77,1.01,1.12,1.56,0.3,0.72,1.02,1,2.22,0.53,1.22,0.56,0.34,2.24,0.33,1.05,0.31,0.73,2.09,1.09,0.7,0.57,0.73,0.33,0.3,0.33,1.73,0.51,0.31,0.57,0.8,0.71,0.32,1.25,0.34,0.32,0.68,1.51,0.52,0.3,0.52,0.32,1.23,1,0.71,0.33,1.58,0.7,1.27,0.37,0.56,1.5,1.52,0.97,0.77,0.72,0.31,0.41,1.02,1.03,1.74,1.01,0.3,0.41,0.7,1.24,1,1,0.6,1.31,0.32,0.7,0.38,1.59,1.01,0.38,0.55,2.02,0.24,0.34,0.9,0.31,0.31,1,0.97,0.4,0.62,1.03,1,0.28,0.48,1.01,1,0.43,1.01,0.38,0.3,0.7,0.91,0.66,1.01,0.32,0.33,1.56,0.79,1.01,0.71,2.01,0.5,1.04,1.21,0.41,1.16,0.32,0.58,0.38,0.34,0.5,0.32,1.15,1.05,0.54,0.3,1.29,0.4,0.33,1.03,0.7,1.2,0.3,0.33,0.5,0.6,0.62,1.07,1.13,0.71,0.32,0.91,1.13,0.9,0.71,0.3,1.69,0.58,1.59,0.28,0.53,0.39,2.16,0.31,1.55,1.6,0.4,1.01,1.03,0.92,0.62,0.3,0.4,0.7,0.9,0.82,0.5,0.92,0.31,0.91,0.4,0.4,0.26,1.11,2.48,0.32,1.09,0.42,2.01,2.02,0.7,1.01,0.32,0.3,0.7,0.38,0.31,2.09,0.41,0.3,0.9,0.7,0.28,0.9,0.54,0.28,0.31,0.93,2.02,0.41,1.37,0.37,2.11,0.23,0.51,1.09,1.18,1.17,0.67,1.6,0.76,1.04,2.01,1.06,1.24,1.06,1.16,1.13,0.51,0.5,1,1.08,1.55,0.41,0.91,0.4,1.07,0.57,1.57,0.71,0.31,0.3,0.35,0.45,0.52,0.38,1.23,0.32,0.51,0.39,0.9,1.18,1.53,0.3,0.76,0.51,0.42,0.41,1.05,0.52,1,0.28,0.23,1.01,0.77,1.51,1.7,1,0.32,1.11,0.85,1,1.08,1.5,0.51,1.5,1.26,1.01,0.31,0.53,0.41,0.53,0.43,0.51,0.8,0.71,0.34,0.9,0.5,1.51,2.01,1.64,0.33,1.04,0.78,0.76,0.52,0.44,1.11,0.9,0.56,0.79,0.71,0.73,0.4,1.03,0.32,0.52,1,1.33,1.54,1.7,1.01,0.57,1.21,0.32,1.01,1.04,0.5,0.32,1,1.3,0.31,0.5,0.43,1.24,0.32,0.53,1.15,1.21,1.04,0.27,1.04,0.71,0.27,0.32,1.5,1.01,1.02,0.56,2.06,1.2,0.73,0.32,0.73,0.9,0.25,1.01,1.09,0.41,0.31,0.53,0.52,0.58,0.38,0.73,0.35,2.04,0.32,0.75,0.5,0.72,0.5,0.32,0.53,0.72,0.79,0.35,0.7,0.59,0.33,1.45,0.3,0.32,0.31,0.45,0.32,0.31,0.7,0.96,1.5,1.01,0.34,1.2,0.43,1.04,1.13,1.22,1.09,1.29,0.4,1,0.39,0.41,0.9,1.01,0.72,1.02,1.02,0.79,2.16,0.37,0.3,0.42,0.41,0.56,1.29,0.31,0.4,0.35,1.22,0.4,0.5,1.01,0.32,0.92,0.5,0.31,0.34,0.7,2.08,0.9,0.34,0.42,0.51,0.45,2.12,0.26,0.32,0.33,0.77,0.32,0.9,1.2,1.03,0.26,0.53,1.01,0.3,0.42,0.3,1,1.01,1.51,0.33,1.31,0.39,0.51,0.9,1.01,0.39,0.32,1.12,0.53,0.5,1.5,0.32,1.45,0.78,0.3,0.4,1,2.1,1.5,1.12,1.26,1.6,0.4,0.31,0.55,1.36,0.81,1.01,2.05,0.7,0.38,1.82,0.5,0.32,1.44,0.52,1,1.07,0.8,0.72,2.03,1.38,0.4,0.74,1,2.01,1.01,1.54,0.26,0.44,0.3,0.31,0.5,0.59,2.05,1.02,0.98,0.4,0.7,0.5,0.37,0.7,0.32,0.4,0.55,0.57,0.46,0.55,0.71,1.14,0.41,1,0.3,1.1,0.5,2.01,0.4,0.34,0.3,0.34,1.51,0.45,1.08,1.13,0.3,0.7,0.34,1.14,1.07,0.43,1.2,0.5,0.41,0.31,0.57,0.93,2.03,1.01,1.07,0.73,0.52,0.53,0.37,0.71,1.12,1.51,0.32,1.87,0.41,0.7,1.09,0.38,1.24,1.2,1.14,0.62,0.7,1.01,1.01,1.1,0.5,1.53,0.31,1.01,0.55,1.5,0.32,1.22,0.31,1.11,1.13,0.53,1.03,1.06,1.04,0.72,1.05,0.3,0.7,1.59,0.34,0.73,0.41,0.81,0.73,1.21,1,1.07,0.52,0.74,0.9,1,1.03,0.41,0.39,1.27,0.33,0.34,0.33,1.22,0.52,0.71,0.31,0.8,2.06,0.51,0.31,0.5,0.32,1.01,1.02,0.34,0.77,0.32,1.21,1.47,0.72,2.42,1.01,0.72,0.33,0.7,0.7,0.4,0.51,0.5,1.59,1.2,0.61,1.01,0.7,1.13,2.02,1.23,0.31,0.71,0.3,1.17,0.32,0.92,0.32,2.19,0.53,0.3,0.77,0.32,0.32,0.3,0.96,0.42,0.35,0.31,0.81,0.53,1.2,2.01,0.41,1.54,0.95,1.61,1.04,0.34,0.3,0.51,0.8,1.17,1,0.51,0.32,0.41,1.1,2.39,1.7,1.28,1,0.52,2.04,1.01,1.01,1.5,0.53,2.01,0.71,0.31,0.34,0.42,0.36,0.3,0.41,0.6,0.33,0.35,0.36,1,1.05,0.41,0.32,0.56,1.18,0.49,1.05,1.11,0.52,1.14,0.36,0.52,1.35,0.5,0.91,0.52,0.62,1.51,0.71,0.4,0.48,0.91,1.57,0.55,1.51,0.4,1.24,0.3,0.32,0.3,0.7,0.34,0.22,1.56,0.53,1.01,0.56,1.25,1.01,0.79,0.43,0.5,1.01,1.59,1.2,0.28,1.09,0.76,0.9,0.4,0.54,1.51,0.77,0.79,0.5,0.23,0.78,0.63,0.91,0.36,0.33,0.31,0.38,0.38,1.21,0.33,2.03,1.01,0.33,2.18,1.54,0.41,0.33,0.7,1.12,0.51,1.16,0.91,1.5,0.3,2.01,0.33,0.55,0.3,0.4,0.71,1.5,0.91,0.5,0.7,0.52,0.39,0.28,1.58,1.34,0.91,0.41,0.33,0.56,1.05,1.03,0.53,0.93,0.31,1,1.26,0.42,1.06,0.38,1.41,0.4,1.51,1.52,1.08,1.5,0.44,0.3,1.27,0.3,1.1,1.54,0.9,0.4,0.35,1.09,0.71,0.57,1.5,2.09,0.3,2.01,0.5,0.41,0.82,0.42,0.7,0.31,0.59,0.55,1.5,1.03,1.01,0.35,0.56,0.7,0.9,0.38,0.72,1.13,0.61,0.32,0.78,1.03,0.23,1.01,0.71,0.37,1.21,0.5,0.53,2.21,0.7,0.45,0.66,0.33,0.52,0.3,2.04,0.36,0.34,1.01,1.5,0.24,0.34,0.71,1.07,1,0.27,2.14,2.24,0.7,1.19,1.5,0.56,0.79,0.77,0.34,1.01,0.3,0.71,1.01,0.34,0.7,0.3,1.03,0.57,1.13,0.71,0.62,0.41,0.3,0.71,0.92,1.24,0.53,1.01,0.4,0.7,1.02,0.59,0.72,2.03,0.57,0.35,1.06,1.21,0.76,0.42,0.4,1.1,0.77,1.2,1.34,0.6,0.77,1.52,1.23,0.72,0.3,0.42,1.52,2.11,0.58,0.36,0.51,0.93,0.64,0.94,1.7,0.25,0.32,0.56,0.95,0.29,0.33,0.41,1,1.02,0.3,2.05,0.7,0.55,1.58,1.01,2.03,0.5,0.32,0.38,1.08,0.38,0.59,0.9,1.03,0.78,1.5,1.54,2.05,0.53,1.24,0.45,0.51,0.25,0.36,0.58,0.51,0.9,0.7,0.3,0.4,0.41,0.31,0.53,0.25,0.55,0.41,0.7,0.73,0.32,0.5,0.33,0.31,1.51,0.8,1.41,0.34,0.41,0.7,0.39,0.29,2.38,0.9,0.36,0.7,0.25,1.01,1.16,1.01,0.7,0.3,0.9,0.4,1.6,0.3,0.9,0.7,0.51,0.3,1,0.55,0.5,0.31,0.57,1.01,1.02,0.5,0.25,0.72,1.23,1.26,0.52,0.7,0.32,1.28,0.7,0.53,0.3,1.88,0.9,1.52,1.4,0.32,1.04,0.9,2.1,2,0.36,0.33,0.32,0.54,0.4,1.09,1.01,1.03,0.78,0.42,0.34,1.06,0.32,0.71,0.31,0.37,0.34,0.7,0.35,0.43,0.5,1,0.9,0.28,0.41,1.18,1.02,1.2,0.74,1.2,0.7,0.53,0.26,0.32,1.15,1.07,0.52,1.04,1.01,0.7,0.56,1.01,0.32,1.11,1,0.51,1.7,1.43,0.55,0.51,1.51,0.74,0.9,0.37,1.01,1.04,0.31,1.01,2.07,0.31,0.31,0.8,0.72,0.82,0.31,0.39,2.08,0.42,0.34,0.36,0.6,0.9,0.74,0.74,1.48,0.33,0.37,0.71,0.95,0.73,0.37,1.5,1.13,0.4,1,0.85,0.51,1.01,1.02,1.54,0.92,0.76,1.04,0.9,0.37,1.02,1,0.3,0.36,0.41,0.71,0.5,0.34,1.74,0.3,0.4,1.2,1.21,0.57,1.12,0.58,0.7,1.07,0.7,0.7,1.7,1.01,0.33,0.52,0.27,0.4,0.42,0.25,0.37,1.14,0.3,0.34,0.36,0.31,0.91,1.17,0.34,0.33,0.3,0.42,0.3,1.01,0.4,0.41,0.3,0.32,1.02,2.01,0.3,0.31,0.7,1.02,1.04,1.1,1.24,0.61,2.14,1.02,0.31,1.15,0.4,0.26,0.34,0.8,1.21,0.33,0.31,1.51,1.34,0.53,1.17,0.3,2,0.3,2.1,0.32,0.5,0.53,0.31,0.8,0.32,2.01,1.11,0.36,0.72,1.25,1.2,0.73,1.07,0.51,0.58,0.73,1.5,0.53,0.7,0.43,1.5,0.52,1.25,2.1,0.31,0.6,1.01,0.31,0.45,0.83,0.5,0.41,2.5,2,0.7,1.13,0.31,1.01,0.83,0.41,0.5,1.51,1.3,0.71,0.3,0.71,0.46,1.03,0.37,2.51,1.07,1,0.7,0.25,0.34,0.3,0.55,0.4,0.51,0.3,0.51,0.42,0.51,1.11,0.4,0.43,0.36,1.16,0.52,1.03,0.33,0.41,0.53,0.27,0.38,0.34,0.41,0.3,0.3,0.46,1.56,0.41,0.8,0.24,1.86,0.57,0.28,1.04,0.32,0.31,1.1,0.9,0.57,0.3,0.66,0.32,0.52,1.34,0.3,1.22,0.3,2.51,1.52,1.01,1,0.34,0.52,0.46,0.52,0.96,0.4,1.62,0.32,1.07,1.74,0.35,1.74,1.01,1.01,0.71,0.75,0.32,0.74,0.71,1.01,2.08,0.34,0.75,1.01,0.31,1.24,0.31,0.69,1.24,0.53,0.24,1.01,1.3,1.09,0.75,0.32,0.7,1.21,0.3,0.71,0.38,0.5,2.02,1.19,0.25,0.33,0.41,0.34,0.34,0.3,0.41,0.93,0.45,0.71,0.38,1.57,0.32,1.55,0.9,1.2,0.76,0.6,0.95,0.73,2.3,0.3,0.3,1.54,0.51,0.55,0.31,0.26,0.9,0.5,1.01,0.93,0.5,0.41,2.01,1.1,0.54,0.41,1.2,0.34,1.7,0.77,1.11,0.9,1.02,2.6,0.83,1.15,0.3,0.34,1.65,1.51,1.26,0.37,0.72,0.47,0.74,0.7,0.31,0.71,0.4,0.3,0.54,0.3,0.32,0.36,0.54,0.32,1.09,1.18,0.35,1.29,0.3,0.41,0.3,0.7,0.37,0.24,0.41,2.04,0.32,0.71,0.3,0.71,0.51,0.56,0.27,0.27,1.01,2.01,0.3,0.54,0.39,0.38,0.91,1.01,0.92,0.62,2.44,1,0.38,0.57,1,1,0.51,0.31,0.94,0.76,1,0.93,0.84,0.34,0.72,0.31,0.23,1.16,0.5,0.5,1.66,1.01,1.58,0.57,1.56,2,0.7,0.4,1.02,0.73,0.32,0.55,1.03,0.41,0.33,1.35,1.51,0.53,1.06,1.2,1.25,1.01,0.51,0.7,0.71,1.03,0.31,0.36,0.36,0.32,0.3,1.29,0.31,1,1.04,2.02,0.42,0.53,0.3,0.53,0.55,0.3,0.3,0.29,0.62,1.05,1.51,0.32,0.62,0.37,0.32,0.3,0.42,0.57,1.51,0.98,0.56,1.27,0.3,1,0.31,0.32,1.27,0.55,0.31,1.16,0.39,0.41,0.42,2,0.55,1.59,0.31,0.9,0.43,0.31,0.9,0.42,0.85,0.23,0.8,0.32,1.01,0.59,0.7,0.27,0.64,1.08,0.26,0.43,0.33,1.23,1.51,0.94,1.23,0.3,0.31,2.19,0.38,0.9,0.34,0.35,0.33,1.51,0.32,0.5,1.01,0.91,1.51,1,1.13,0.5,0.8,0.36,0.36,1.25,1.58,0.32,0.3,0.8,0.39,0.42,0.42,0.7,0.54,0.42,0.74,1.02,0.31,0.36,2.02,0.91,1.57,0.33,1.05,1.01,0.51,1.02,0.7,1.18,1.12,0.32,2.13,0.3,1.02,0.38,0.72,2.01,0.41,1.25,1.04,1.18,1,0.72,1.2,0.55,1,1.05,1.51,0.31,0.75,0.56,1.01,0.41,0.7,1.01,0.71,1.2,1,1.51,0.9,1.01,0.43,0.46,1.5,0.41,0.42,1.51,0.5,1.02,0.75,0.53,0.8,0.7,1.01,0.9,0.61,0.55,0.31,0.52,0.31,0.74,0.41,0.31,0.56,0.36,0.5,1.8,0.71,0.52,1.01,0.53,2,2.16,0.53,0.32,0.36,0.84,0.7,0.31,0.7,0.4,1.02,1,1.33,1,1.08,0.53,0.3,0.5,1.01,0.8,0.54,0.81,0.56,1.01,0.32,0.54,0.3,0.3,0.4,1.01,1.17,0.35,0.32,1.51,0.4,1.6,0.31,0.31,0.99,2.01,0.27,0.3,0.7,0.67,0.24,0.71,0.54,0.71,2.09,0.32,1.01,1.06,0.7,0.3,2.1,0.51,0.54,1.03,1.51,0.34,0.41,0.46,1.04,0.33,0.51,0.34,0.31,1.23,0.33,0.34,0.31,0.65,1.58,0.23,2.02,1.83,0.91,0.31,1.02,1,0.36,1.51,1.34,0.92,1.16,0.3,1.01,2.15,0.9,0.9,1.33,0.54,0.33,0.31,1.2,0.41,0.51,1.85,0.32,1.32,0.76,0.23,0.54,0.32,1.02,0.5,1.02,0.44,1.01,1.23,0.9,1.53,1.3,0.73,0.31,0.34,0.27,1.63,0.5,0.74,1.16,0.3,2,0.3,1.32,0.56,0.38,0.32,1.12,1.2,0.53,0.37,1.61,0.79,1.51,1.04,1.01,1.53,0.3,0.3,1.03,0.3,1.05,0.31,0.3,0.31,0.36,0.51,0.75,0.71,0.38,0.43,1.06,0.72,1.25,0.9,0.53,1.21,0.83,1.52,0.3,1.5,1.21,0.77,0.39,0.4,0.32,0.92,0.36,1.02,0.25,0.3,0.3,1.5,1.01,1.01,1.06,0.9,0.7,0.5,1.59,1.18,1.11,1.04,0.74,0.28,1.08,0.52,0.36,1.03,0.36,2,0.7,0.77,0.7,0.31,0.35,0.42,0.67,0.56,2,0.9,0.42,0.33,0.6,0.41,1.01,0.83,0.3,0.34,0.35,0.76,0.9,2,1.51,0.7,0.3,0.7,1.01,1.13,1.52,0.4,0.31,1.09,0.52,0.39,1.39,0.41,0.7,1.01,1.1,0.32,0.35,0.31,0.3,1.52,0.9,0.37,0.71,1.64,0.59,1.09,2.01,0.41,1.26,0.76,0.35,0.7,1.51,0.28,0.9,0.96,1.03,1.51,1.1,1.09,1.03,1.06,0.31,1.53,0.52,0.7,1.5,1,1.07,0.31,0.33,0.4,0.32,0.91,0.5,0.32,0.37,1.1,0.34,0.51,0.4,1.14,1.05,0.32,1,0.51,1.02,0.32,0.32,0.3,0.71,1.01,0.39,1.52,0.44,0.54,0.72,1.01,1.04,0.9,0.3,1.7,1.01,0.93,0.5,0.25,0.3,0.71,0.41,1.31,1.16,0.71,0.3,1,1.13,0.34,0.68,0.4,0.53,0.7,0.41,1.61,1.21,0.32,0.63,1.13,0.71,0.23,1.01,0.81,1,1.22,1.53,0.32,0.33,0.39,1.08,0.92,0.72,0.54,0.73,0.36,0.71,0.7,0.34,0.41,0.72,0.31,1,1.77,0.9,2.01,0.54,1.21,1.7,0.44,0.9,1.22,1.55,0.81,0.5,1.3,0.7,1.02,0.5,0.33,0.41,0.76,0.61,1.2,0.58,0.27,1.2,1.01,0.51,0.31,0.4,0.71,1.56,0.41,0.4,2.36,1.55,0.24,0.51,1.1,1.02,1,0.5,1.5,0.73,0.31,0.56,0.38,0.54,0.38,0.77,0.51,0.73,0.71,0.42,1.51,0.52,1.55,0.76,1.01,0.73,0.91,0.39,0.33,0.37,1.01,0.41,1.12,1.15,0.42,0.42,0.3,0.38,1.2,0.42,0.7,0.71,2,0.48,0.29,0.38,0.9,1.3,0.33,2.52,0.33,0.62,0.31,0.54,1.04,0.4,0.42,0.51,0.92,0.5,0.41,0.7,0.7,1.59,0.3,1.51,1.04,0.31,1.05,1,2.01,0.55,0.71,0.23,1.5,0.71,0.71,2,0.7,0.79,2.28,0.37,1.27,0.36,1,0.82,0.42,0.57,1.26,1.39,1,1.01,1.15,0.33,0.24,0.7,1.34,0.37,0.31,2.2,1.08,1,0.52,1.01,1,0.75,0.41,0.24,1.5,1.51,0.8,1,0.54,1.07,1.08,1.5,0.41,1.5,0.7,0.37,0.8,1.02,0.5,0.78,0.31,1.06,0.5,1.15,0.25,2.26,1.05,0.57,1.01,0.32,0.53,0.91,0.41,1.23,1.31,0.9,0.42,1.13,0.34,1.05,0.58,0.3,1,0.42,0.91,0.32,0.86,1.33,1.52,0.72,0.7,0.53,0.5,0.42,1.03,1.16,2.04,0.4,1.01,0.5,1.04,0.75,1.26,1.06,0.34,0.32,1.01,0.91,0.36,0.9,1.2,0.9,1.23,0.75,0.5,0.53,0.3,0.91,0.51,2,0.75,0.9,1.42,0.34,1.3,1.51,1.01,1.27,2.01,0.4,0.27,1.03,0.73,2.02,0.38,0.36,1.51,1,1.51,0.32,1.09,1.34,0.4,1.04,0.7,0.3,0.31,0.76,0.27,0.33,0.64,0.39,0.7,1.02,1,0.72,0.7,1.23,0.74,0.3,0.42,0.71,0.76,0.38,0.9,1.02,1.67,0.51,0.9,1.2,0.9,1.53,1.11,0.41,1.32,0.38,0.35,1.52,0.9,0.24,0.75,0.91,0.4,0.33,1.31,0.36,2.01,0.33,1.51,1.25,1.02,1.22,0.34,0.33,0.7,0.77,0.5,0.31,0.3,0.56,2.02,1.02,1.01,1.51,2.04,0.71,1.57,0.35,0.53,0.32,0.78,0.53,2.1,0.82,0.71,0.3,1.05,0.7,0.33,2.01,2.01,1.01,0.42,1.02,0.93,0.81,2.61,1.18,0.7,0.28,0.39,0.71,1.59,1.04,0.53,0.9,0.32,1.18,0.31,0.31,0.3,0.4,1.54,1.11,0.57,1.5,1.65,0.72,0.29,0.52,0.76,1.06,1.62,0.71,1.21,0.52,1.12,0.96,1.01,0.3,0.32,1.51,0.3,1.16,0.66,0.79,0.59,0.57,1.01,1.26,0.44,0.5,1.5,0.51,1.2,0.71,0.9,1.06,1.24,0.53,0.72,1.01,0.36,1.14,0.3,1.58,1.11,1.21,0.3,0.33,0.81,0.39,0.51,1.01,0.42,0.39,0.92,1,1.57,0.32,0.91,0.44,0.53,0.58,0.32,0.7,0.38,0.3,0.7,0.77,2.01,0.71,1.71,0.42,2.06,1.98,1.01,0.54,0.5,1.04,0.36,0.61,0.24,0.91,1,0.31,0.31,1.73,0.3,0.31,0.42,1.01,0.52,1,1.27,1.22,0.54,0.31,0.98,0.84,0.31,0.33,0.3,2.5,1.52,0.38,0.75,0.43,1.02,0.77,0.35,0.33,0.5,1.02,0.72,0.41,0.38,0.7,0.41,1.01,0.31,1.08,2.02,1.56,0.73,1.01,0.8,0.35,0.71,0.41,0.86,0.9,1.46,0.4,1.22,0.53,0.71,1.52,0.3,0.42,0.42,1.41,0.3,1.51,0.5,1.68,1.53,0.54,1.14,2.4,0.42,1.03,0.9,1.05,1,0.41,1,0.7,0.3,0.54,1.01,0.71,0.7,1.01,0.4,0.58,0.38,1.08,0.32,0.34,0.3,0.32,1.51,0.33,0.71,0.48,1.09,0.72,1.67,0.75,0.27,0.41,0.55,0.46,0.34,0.7,0.54,1.01,2,0.4,1.07,0.51,0.51,1.51,1.01,1.02,0.31,0.3,0.3,1,0.36,0.51,1.02,0.7,0.58,0.33,1.01,1,0.76,0.3,2.01,0.9,0.35,0.52,0.34,1.03,1.03,0.29,0.36,0.42,0.3,0.55,0.5,1,0.31,0.72,0.54,0.3,1.56,0.9,1.11,0.83,0.28,0.43,0.55,0.33,1.02,1.01,0.57,1.01,0.55,0.33,0.9,0.33,0.54,0.41,1.15,1.03,0.41,0.59,0.31,1.62,0.3,1.37,1.01,0.31,0.32,0.58,1.54,1.13,0.8,0.4,0.96,0.56,1,0.5,0.3,0.51,1.5,1.01,0.58,0.53,0.38,0.56,0.28,0.53,0.86,0.59,0.27,0.73,0.6,0.35,0.57,1.5,1.13,1.51,0.81,0.23,0.3,0.78,0.25,1.5,0.93,0.52,1.23,0.73,0.32,0.72,1.5,1.12,1.5,0.42,0.3,0.34,0.92,0.43,1.1,0.87,0.96,0.71,1.23,0.24,0.6,0.7,0.3,1.2,0.34,1.6,1.52,1.09,1.2,1.59,0.24,0.41,0.54,1.31,0.32,0.35,0.31,1,1.95,0.32,0.4,0.71,0.7,1.15,1.21,0.52,0.7,0.82,0.26,0.92,1.11,0.43,2.2,1.38,0.51,1.01,1.01,2.03,0.52,1.21,0.44,1,1.04,1.29,0.41,1.08,0.71,1.65,0.52,1.11,0.32,0.74,0.3,0.3,0.41,0.63,1,0.7,1.16,0.4,0.69,1.01,0.33,0.5,1.79,1.05,1.09,0.79,0.51,0.35,1.02,1.06,1.01,0.5,1.51,0.3,0.31,1.2,1.03,0.41,1.11,0.33,0.54,0.32,1.26,0.46,0.44,0.33,0.58,0.3,0.5,1.15,0.76,1.5,0.51,2,0.72,1.12,0.7,0.7,0.32,1.04,0.81,0.44,0.38,0.32,2.03,0.39,1.34,0.77,0.51,0.33,0.9,1.53,1,0.38,1,1,1.7,0.55,1.13,0.57,0.31,0.41,0.9,0.32,0.55,0.91,1.5,0.33,0.35,1.5,1.01,0.3,0.34,0.33,1.01,1.04,0.41,0.71,1.05,1.07,2.02,0.34,0.73,0.71,1.24,0.26,0.31,0.38,0.34,1.2,0.5,0.42,1.75,0.61,0.36,0.53,0.9,1.36,0.24,1.23,0.31,1.01,0.36,0.3,1.05,0.32,2.02,1.21,0.65,0.7,0.41,0.43,0.41,0.4,0.55,0.52,1.02,1.02,0.35,0.31,1.03,0.72,0.41,0.71,0.47,0.31,2.01,0.5,0.34,1.23,1.14,1.53,1.03,1.71,1.33,1.04,0.3,0.33,1,0.3,1.02,0.54,0.83,1.05,0.31,0.32,2.04,0.73,1.01,1,0.3,0.44,0.53,1.71,1.02,0.61,0.71,1.41,1.11,0.52,0.74,0.9,0.31,0.32,1,0.7,1,0.72,0.4,1.59,0.83,1.61,0.3,1.26,0.56,0.38,1.12,0.74,0.31,0.42,0.83,0.5,0.81,0.71,0.31,0.7,0.53,0.33,0.31,1.17,1.12,0.5,1.21,0.33,0.32,0.73,1.04,0.31,0.23,0.3,1.01,1,0.54,1.05,0.35,0.56,0.3,0.32,0.68,0.4,1.52,0.89,0.32,2.4,0.82,0.27,0.55,1,1.02,0.6,0.31,0.36,1.53,0.32,1.54,0.72,0.59,0.31,0.7,0.33,0.72,0.71,0.3,1.13,0.42,0.7,0.31,2.06,0.7,0.33,1.08,0.9,0.56,0.3,1.5,0.31,1.02,2.06,1.19,2,0.74,0.46,0.5,0.34,1.53,0.71,2.65,1.29,0.57,2.54,1.59,0.67,1.01,0.5,0.32,1,0.35,1.34,0.72,0.8,0.41,2.01,1.03,1.5,1,0.31,0.37,1,0.39,1.22,0.56,0.24,0.7,0.9,0.26,0.32,0.9,0.75,0.32,2.01,1.13,1.01,0.55,0.31,1.05,0.76,1,1.11,0.3,0.7,0.34,0.55,0.32,0.24,1.5,0.81,0.31,0.94,1.21,1.2,0.3,0.7,0.92,2.35,1.79,2.01,0.61,0.34,1.15,0.5,0.42,2.02,1.5,0.83,1.36,1,1,1.56,0.3,1.06,2.06,0.32,1.16,0.72,0.58,0.23,0.74,0.51,0.44,0.33,1.38,2.02,0.4,0.32,1.01,0.59,0.61,1.19,1.04,0.23,0.59,1.02,0.53,0.71,0.55,0.44,0.39,1.34,0.31,0.56,1,0.31,0.42,2.01,0.71,0.91,1.59,1.15,0.5,1.5,2.1,0.63,1.51,0.52,0.32,1.08,1.01,1.19,0.52,2.2,0.32,1.28,0.3,1.61,0.32,1.52,0.49,1.01,1.17,0.52,0.74,0.91,0.7,1,0.4,0.39,0.51,0.35,1,0.55,0.38,0.92,1.2,1.01,1.02,0.51,0.4,1.14,1.3,1,0.57,1.16,1.5,1.03,0.35,0.43,0.9,0.76,0.51,0.55,0.36,0.91,0.32,0.71,0.26,0.3,0.26,1.2,0.32,3.02,1.27,0.73,1.52,1.06,0.32,1.6,0.6,1.01,2.51,0.35,0.4,0.33,1.09,1.01,1.02,0.9,0.57,0.4,1.17,0.34,1.53,2.07,0.36,0.31,0.7,0.32,0.54,0.71,2.01,0.59,0.54,0.32,1.01,0.24,0.33,0.32,0.58,0.6,0.33,0.7,0.46,0.88,1.01,1.02,1.02,0.56,1.14,0.97,0.71,0.55,0.38,1.22,0.29,0.26,0.7,0.42,1.17,1.29,1.64,0.73,0.41,0.56,0.44,0.77,1.5,1.51,0.5,1.69,2.01,1.21,0.53,1,0.38,0.42,0.7,1.06,0.75,0.52,1.59,1.51,0.3,0.35,0.4,0.37,0.3,0.35,0.4,1,2.01,0.79,0.73,1.42,0.71,0.85,1.26,0.38,1.06,0.33,0.54,1.01,1.21,1.02,1.04,0.51,0.42,1.83,0.51,0.41,0.42,1,0.3,1.14,0.72,0.28,1.35,0.34,0.71,0.9,1.51,0.53,0.78,0.31,1.13,0.81,0.9,0.59,0.32,0.4,0.33,0.78,1.55,0.9,0.72,0.57,0.34,1.5,0.42,0.4,0.41,1.06,0.5,0.77,0.57,0.8,2.14,0.3,0.58,1.5,1.06,0.43,1.01,0.41,0.3,1.01,1.5,0.61,1.52,0.92,1.52,1.01,1.02,2.03,0.34,2.22,0.75,0.71,0.32,0.7,1.27,0.44,0.36,0.3,0.73,1.1,1.24,0.35,0.44,0.43,0.3,0.55,0.36,0.7,1,1.11,0.61,1.5,1.24,0.51,1.16,0.42,0.71,0.33,0.3,0.31,1.3,0.55,0.72,0.56,0.4,0.52,0.3,1.02,0.4,0.71,2.09,1.2,0.58,0.9,2,0.54,1.2,1.02,0.31,0.71,0.34,0.24,0.52,0.78,1,0.73,1.51,0.71,1.7,1.21,1.01,0.91,1.01,0.83,0.36,0.3,0.85,0.23,0.31,0.37,1.31,0.34,0.32,0.4,1.01,0.5,2.08,0.36,0.74,2.01,0.33,0.42,0.38,1.02,0.4,2,0.7,0.81,0.33,0.76,0.34,1.5,1.5,2.1,0.92,1.24,1.01,0.92,1.46,0.5,0.7,0.33,0.34,0.52,1.01,1.59,0.55,0.31,0.92,0.31,0.71,0.3,0.39,1.18,1.12,1.21,0.42,2.01,1.52,0.31,0.93,0.28,1.01,0.33,0.38,2,0.33,0.4,1.01,1.01,0.74,0.37,1.53,0.4,0.37,0.56,0.4,1.32,0.5,0.3,0.81,0.9,0.9,1.01,1.6,0.33,1.2,0.7,0.78,0.3,0.31,0.32,0.53,0.38,1.01,0.32,1.55,0.73,1.51,1.5,1.03,1.03,0.53,0.72,0.27,0.67,0.34,0.57,0.23,1.05,0.33,0.71,0.32,1.01,0.74,0.52,1.13,0.75,1.42,0.9,0.32,1.01,1.16,0.71,1.52,0.51,1.11,2,0.52,0.78,1,0.78,0.75,0.31,0.34,0.7,0.3,0.3,0.9,1.03,1.27,0.33,1.2,0.34,1.05,0.6,0.48,0.48,1.07,0.32,0.3,1.21,0.7,1.12,0.76,1.28,1.14,1.74,1.09,1.82,1.13,0.32,0.51,0.5,0.71,0.27,1.01,0.3,0.7,1.08,1.22,1.13,0.51,1.25,0.3,0.47,0.72,0.72,0.4,2.02,0.43,0.36,1.06,0.7,0.74,0.51,1.06,0.5,1.51,0.35,0.91,0.53,0.52,0.25,0.59,0.72,0.7,0.35,1.08,0.79,0.5,1.02,0.26,0.73,0.31,0.72,0.31,0.4,0.72,2.01,0.75,1.53,0.33,1.01,0.7,0.51,0.9,1.12,0.94,0.71,0.54,0.31,2.28,1.04,0.31,0.5,1,1,0.74,1.02,0.41,1.78,0.4,0.73,1.07,0.41,0.43,0.71,0.59,0.74,0.71,1,1.02,0.51,0.43,0.32,1.58,1.51,1.52,0.41,1.01,0.33,1.13,1.29,0.4,0.76,0.43,0.71,0.34,0.26,0.31,0.35,0.4,0.27,0.33,1.62,1.05,1.01,0.96,0.33,1.73,1.21,0.51,1.29,1.24,0.8,2.01,0.76,0.74,1,0.52,0.71,1.09,1.37,1.01,0.3,1.01,2.03,0.31,0.9,0.36,0.37,0.32,1,0.77,0.32,0.4,0.32,1.09,1.05,0.3,0.3,0.72,0.8,0.7,1.05,0.32,0.93,0.73,1.01,0.75,0.72,0.76,2.01,1.02,2.06,0.51,0.35,0.3,0.39,0.35,1.01,0.52,0.32,0.55,0.24,0.9,0.41,0.4,0.32,0.5,1,1.23,1.3,0.5,0.7,0.61,0.28,0.92,0.59,0.27,1,1.52,0.7,0.59,2.04,0.52,1.08,0.34,0.5,0.33,0.71,0.32,1.77,0.3,1.01,1.53,1.7,0.53,1.15,1.01,0.41,0.41,0.82,0.53,0.26,0.7,1,1.01,0.41,1.05,0.9,1.01,1.01,0.3,0.73,0.39,1.19,0.55,0.38,0.71,0.71,0.3,1.07,0.63,1.5,0.6,0.32,0.3,0.43,0.25,1.5,1.36,1.21,0.31,0.34,0.43,0.98,1.09,1.22,0.81,0.33,1.04,0.9,0.3,2.06,1.2,0.57,0.34,0.96,0.31,1.7,0.5,0.7,0.63,0.46,1.1,1.54,1.52,0.52,0.51,1.31,0.38,0.42,0.91,1,0.44,0.8,0.37,0.42,1.04,0.34,1.51,0.31,1.71,0.25,1.16,0.32,1.52,1.01,0.3,0.34,1.31,1.21,0.31,0.36,0.33,0.39,0.55,0.43,0.32,0.41,1.03,1.55,0.35,1.5,1.09,0.57,0.5,0.53,0.31,0.32,1.61,0.9,0.7,0.5,0.33,0.32,0.56,0.33,0.42,0.38,0.76,0.91,1.11,1,0.51,1.17,1.06,0.33,0.58,1.38,0.31,1.2,0.26,0.3,1.32,0.3,0.23,1,0.33,1.01,0.54,0.42,0.72,0.9,0.31,0.42,0.7,0.51,2.02,0.9,0.91,1.22,0.3,1.51,1.13,0.33,0.9,1.01,0.75,1.2,0.91,0.7,1.53,1.02,0.52,0.52,0.92,3.01,0.33,1.23,0.26,0.3,0.58,1.21,0.31,0.22,1.22,1.58,0.5,0.5,1.2,0.41,0.31,1.51,0.51,1.01,0.37,0.4,1.51,1.39,1.11,1.22,0.35,0.27,0.3,1,1.02,0.3,1,1.2,0.77,0.35,1.09,0.8,1.23,1.01,0.36,2.22,0.75,1.02,0.93,0.76,0.33,0.57,0.31,0.62,0.7,0.5,0.42,0.31,0.55,0.41,0.47,1.09,0.33,0.71,0.32,0.91,0.9,0.36,0.71,2.01,1.51,0.32,0.55,1.02,0.7,1.52,0.3,1.96,0.39,1.32,0.71,1.23,1,1.3,1.57,1.25,0.65,0.41,1.02,1.2,0.58,0.57,0.91,1.09,1.52,0.28,0.81,2.02,0.41,1.57,0.74,0.43,0.33,0.32,0.74,1.37,0.58,0.9,0.9,1.4,0.54,0.92,0.71,1.22,1.2,1.4,0.71,0.4,0.3,1.31,0.41,0.41,0.36,1.06,0.56,1.02,1,0.77,1.03,1.57,0.37,2.17,0.67,0.27,1.21,1.52,0.3,0.31,1.5,0.32,0.31,0.4,1.47,1.51],"y":[2397,3300,713,707,987,3250,1668,1771,1053,4640,2467,758,5755,6097,17422,12343,4525,734,685,4326,5546,2100,12829,4002,7274,698,2643,683,1207,403,5824,1116,4200,1441,545,673,5013,1351,1374,5219,3985,829,9882,1103,4639,8312,4579,16137,9173,5817,3950,1656,534,1400,2386,682,16733,596,6089,3748,532,802,6456,14388,5761,5783,11200,12845,472,5852,6975,596,2042,1143,875,1013,8818,6969,766,965,9243,1153,734,1395,988,840,7747,4974,6619,1569,1721,16670,523,15031,953,662,3472,645,2548,1412,1232,1863,1881,5868,7056,2376,16116,1000,10781,460,555,4853,1944,734,1323,4508,5937,15110,5958,514,4081,2686,4113,2501,2839,1901,765,2827,789,731,3429,2546,6250,8057,1745,3142,3160,7667,453,1268,4296,5938,12485,14199,1173,4844,18108,5292,8637,2367,2926,768,10494,710,5599,2172,2257,7533,5622,8073,2381,607,4084,4508,999,702,3819,1607,459,5281,9256,3061,1289,9271,1187,14196,478,1116,776,4816,383,4872,1771,2751,1559,10962,3468,538,4363,675,4871,7145,1662,2616,2125,692,402,1749,2656,1643,4256,1591,3664,526,3355,2391,12185,3787,5056,814,628,3819,15092,4969,2266,2670,12008,3189,1599,3026,8408,1642,2686,1974,7277,3959,7350,13853,5516,1030,875,558,5450,1356,2642,648,1775,2936,783,2648,12265,4689,1436,1015,3512,4309,10147,3114,523,9090,2528,12809,4031,2353,2113,3282,638,11975,8501,1903,5821,2092,647,2500,13853,1184,3869,493,972,3448,11021,2576,1792,3724,4760,596,854,7560,1031,5468,698,1811,926,4956,6652,393,4544,1016,4025,11360,612,1224,13015,663,854,1899,13786,2788,579,1820,2130,1116,2992,8973,554,4653,1062,470,3634,6539,3676,781,419,6204,368,4594,17153,1354,552,2590,6563,429,2596,1359,723,2940,906,781,645,619,6786,12117,3312,758,1438,1687,4535,4381,956,625,3734,3402,1935,2282,1841,984,1378,576,1953,4306,6885,2838,4427,1879,1813,675,4309,3253,738,7145,4238,919,4939,871,1562,1669,1225,4657,8145,5893,2135,914,2157,13445,548,906,1018,828,3674,3382,999,878,9157,3249,1627,938,2375,419,1060,593,3002,784,646,4826,10428,8475,4742,3278,3519,657,702,4543,979,4269,760,3307,7114,1282,2817,18118,526,1193,540,418,668,5919,15649,14561,4173,1052,758,622,684,10395,821,2291,2732,7438,2442,14959,9095,710,941,907,4429,919,949,698,1833,854,2700,2871,898,7333,6383,1814,4989,12451,1799,4492,957,2516,1206,3089,11322,4972,17247,7182,3082,765,3774,4416,2139,499,5423,1883,2020,873,507,14527,6405,480,5292,8868,3465,976,965,974,540,7741,6793,16256,16297,1200,4528,1069,638,5610,5559,6333,3685,7695,15841,2686,645,393,5622,4480,2398,489,941,2229,5331,4189,13853,1821,4766,905,462,2652,2536,855,3231,5967,877,7375,1035,3266,1743,625,9257,683,7440,3250,530,828,554,608,1002,1125,4630,3589,1869,969,936,1172,644,2129,7306,921,840,867,8430,1928,978,776,941,4498,4068,12146,6221,579,610,3145,10165,11390,13782,787,1268,9209,1140,4949,2051,884,713,6532,505,1063,2801,3015,755,779,593,675,8214,1024,5070,596,1122,1115,11255,3734,5226,6167,1591,13068,2534,734,6232,6075,801,2145,4811,1909,8818,628,1011,4064,675,623,2516,13540,844,2753,802,2106,686,613,1949,4265,548,1865,4286,2051,5242,4514,727,3590,15116,1870,4212,3336,18159,2857,2222,2636,5443,678,6075,943,669,1061,5889,18257,13387,5269,2874,667,2879,18026,5818,1686,17366,6542,6104,756,2290,933,7368,656,3906,15092,9092,7617,8815,5845,4899,3050,11512,2040,670,459,3839,4124,1196,14483,10886,1050,3734,5063,7988,4365,3304,584,741,1911,14740,10688,6047,3005,2208,5204,814,1359,6733,2486,4068,530,13603,854,2369,3184,1273,15618,14433,7677,478,2030,1372,1235,7633,898,9659,3167,1061,3833,1348,5219,3862,1815,1053,1128,3555,833,2673,13068,8879,2338,1365,5376,698,1076,2604,2804,2811,2357,12338,1170,803,15769,2932,796,10706,1259,1076,716,421,449,7190,1121,3653,12308,3163,9836,828,511,3360,1015,847,1062,1655,734,701,2799,16409,2264,805,5581,8972,8921,2331,766,2314,2898,607,622,3672,1008,10043,1662,5908,1554,12418,4513,10804,4583,6429,2371,642,1078,1945,945,886,7257,926,1883,492,967,1202,574,1637,4623,776,802,13162,3713,435,3192,4811,2573,2112,7781,1442,1367,6738,2198,7070,752,2307,4732,608,5671,11577,3284,482,906,1316,9253,5226,720,5522,588,705,6578,4872,4751,2093,698,4602,838,1132,3437,5869,7049,1103,5265,6352,2784,680,5372,6563,1002,1897,1003,5894,1956,2026,5007,540,5420,763,2715,4838,583,2368,1552,2847,7125,15662,544,14931,3347,1694,4167,1273,8951,8313,3045,537,17659,3613,11177,1824,5505,7487,790,5818,3011,1410,5793,3012,862,13215,4670,957,842,5102,3360,4064,14092,1758,4815,8371,645,4357,1744,433,700,432,3368,2431,1402,733,3063,2041,2037,4973,2348,1045,4403,529,2935,1067,1760,7553,1554,4168,4106,1263,421,1229,407,2976,9642,5160,1318,734,6426,1358,2348,7806,540,6201,10351,4379,2956,6770,8794,3101,707,3363,811,4946,4196,911,18565,6261,1158,5250,394,9478,2802,1248,1419,1030,872,1708,6079,3775,936,5638,1816,3697,4749,3700,12082,731,1907,3601,6245,18706,1803,6887,1427,984,16709,743,6486,734,2826,13092,4538,2453,1785,2846,743,911,492,8816,1383,732,1573,2862,2735,672,10285,641,796,2074,14465,2012,878,1577,548,6403,3465,3431,1010,10345,2290,6654,703,1091,5410,10308,4063,3321,2708,1034,791,4924,9205,8731,6666,552,1187,1758,6867,8020,4569,1433,8922,730,2381,871,7123,6244,1193,1238,9728,504,596,3752,921,805,3965,2357,591,2161,5330,4737,646,960,5288,6115,670,8688,879,709,3107,3751,2178,4038,589,965,13787,2482,4118,2320,8104,1154,4770,5190,830,10082,645,1984,1026,413,1706,828,4877,3293,2041,447,4045,1125,723,5050,2512,6814,573,557,1095,1777,1378,7222,5062,1626,905,4392,4873,4580,3072,691,11086,1899,9094,487,1857,1213,17891,802,10546,8138,971,4129,4900,4281,2321,432,737,2150,5141,2893,2475,4096,872,4279,931,684,724,7758,12883,645,7487,992,11825,15974,3585,8716,533,384,2839,733,732,15773,1356,945,3704,1092,642,4435,1786,642,635,5729,13109,964,12455,833,16336,530,1717,6852,5412,7155,3178,16240,2036,5561,9793,2239,13178,4060,4657,7016,1682,1103,3276,4953,14386,1367,3800,954,7577,2753,15144,1917,675,421,503,1817,1737,1064,6206,576,1438,587,3989,12175,13629,886,3549,1569,737,1153,5352,2359,3790,598,364,5215,3345,6851,13949,3752,706,4578,3084,6037,5256,8732,1402,10584,7567,4506,571,1607,1187,1395,795,1436,3296,3123,650,4693,1845,9741,18607,5624,694,3832,3105,4229,1833,1238,4973,3407,1819,4570,3135,2426,667,9691,505,1668,3998,6316,7589,15568,3801,1949,5124,494,3399,5105,1791,864,4389,6443,593,1583,975,10388,648,1607,5750,4824,6278,518,9882,2590,583,808,6340,4513,3987,1580,14982,6026,2326,828,2752,3205,558,3895,10314,1259,625,2158,1644,2227,855,2564,906,9905,648,2588,1094,3274,1190,645,2563,2819,3710,1024,1806,1561,984,10800,878,828,489,1193,972,1074,2048,2801,15992,4475,819,6547,1408,7632,10396,8485,7583,5996,737,4620,1095,648,4068,6806,2578,4044,5312,2898,8709,957,844,1142,827,2298,6588,571,1059,706,5602,890,2645,4194,499,4379,1098,1012,650,2247,17916,3669,775,1142,1343,978,17694,453,730,521,2642,1080,6892,4516,5018,599,1665,7665,666,1096,814,5775,6335,5919,666,6676,958,1146,7747,4559,1008,471,5046,1332,2033,14527,1170,6455,3298,394,868,3789,12121,6250,8721,6566,12467,975,593,1186,5103,2655,4875,15109,2264,1211,12081,1154,645,7580,1767,3562,2375,2753,3614,16945,13250,982,2870,3472,13940,4525,8848,597,990,673,732,1960,1622,15857,5241,4308,900,2338,1890,684,2924,758,687,1556,1963,1314,2224,4007,5291,791,7392,526,3942,1125,14768,951,1174,764,1001,7891,1154,6168,4959,487,1894,517,11654,5393,774,9609,1624,739,802,1645,4214,18578,5919,4686,2330,1147,1428,811,2271,10211,10113,540,17761,1115,2726,4584,566,8069,5458,5925,3968,2373,3852,4578,4827,1746,9596,802,7001,1584,7515,828,7372,462,4771,4936,1648,4292,5143,3603,3478,4797,526,3039,8829,745,3758,994,2797,3311,7703,4389,3296,1749,2906,3348,4312,6056,999,651,10389,814,686,743,5960,1778,2891,891,2550,11854,1875,1046,1935,828,5443,4718,765,3291,972,8286,6108,2567,16198,5786,3236,579,2753,2239,917,1882,1074,9972,5268,1752,4887,2473,6391,15334,8541,628,2790,491,5025,612,4482,702,13978,2177,524,2949,1171,561,895,4187,803,984,335,3097,2137,3505,17334,1007,7287,3750,10197,6564,596,524,1656,2856,4914,3304,1656,846,1192,4497,17920,7414,8895,4620,1600,18795,4559,2828,6750,1939,16677,2233,802,540,1087,518,675,1151,2316,521,1063,794,3640,5236,1015,421,2010,4746,1011,5468,6982,1689,7275,726,1170,4423,1138,3180,1689,1616,8400,3340,1035,994,4332,10964,1799,11666,1066,11376,844,628,545,3214,765,404,8107,1607,5174,1743,5975,4642,2878,1056,1090,4298,7943,5600,707,5127,1896,4536,596,1981,13080,3089,2507,1316,400,2496,1105,3340,810,917,408,815,1176,5882,631,15717,6499,666,16690,7486,932,434,3177,9214,3920,5002,3181,12821,1013,15562,666,1702,731,765,3464,12754,3723,1939,1867,1111,1154,825,11526,5245,2854,827,854,1951,5557,4038,2143,4793,1214,6518,10302,984,8093,855,8686,1038,14247,12071,5044,13653,1003,505,7849,684,8932,10164,3924,877,522,6849,2950,2052,12000,11576,473,15977,1690,1243,3795,984,2553,897,1709,1848,8662,5523,4588,906,1737,2498,3062,633,2949,4766,1805,561,2694,4414,530,5010,2508,957,7513,1745,1813,16558,3011,1027,2618,492,1720,456,18127,624,616,5939,6838,478,517,2863,5532,4788,496,16390,15852,2537,10436,10406,1993,3436,2753,596,4412,810,2162,4751,714,1758,673,4764,1982,11051,3002,1348,918,605,2313,3744,5412,1163,4659,654,3454,7357,1552,4228,12714,1292,879,3996,5324,3183,1042,552,3604,1940,5242,7893,1897,2089,12916,10646,2430,878,945,14217,17871,1865,568,1343,5171,2420,3518,15797,576,477,1924,4738,674,1240,969,4144,4832,515,7026,2854,1744,10546,4171,12862,1433,730,933,5455,1117,1698,2761,10607,2777,9408,12196,13543,2280,6319,941,1443,548,713,1666,1656,3377,2043,475,1008,738,942,1956,575,1272,1115,2352,3411,657,2025,727,707,6756,2329,8654,765,844,2889,1008,669,16643,4309,616,2936,438,4129,5164,4189,2484,473,3740,945,6899,414,3473,1949,1752,382,5088,1081,1240,865,2008,4497,4281,1333,438,2949,6459,7910,1712,2874,708,6726,2453,856,844,8048,3398,7899,8624,494,4164,3324,12381,17869,537,743,645,1662,720,2708,5430,3530,2200,810,805,5293,427,2764,593,1041,880,2100,583,1329,1654,7343,3307,646,930,9137,4037,10454,2327,5655,3296,1564,456,773,9025,3296,1619,4484,6097,2826,1340,3888,905,7179,8888,1268,14611,6727,2303,1268,5979,3030,4297,681,6108,3914,816,6105,16337,872,462,3173,2328,3593,1046,697,16854,756,764,782,806,3464,3180,2410,7086,652,1082,2347,3669,3937,681,14179,10228,790,7507,2442,1800,6239,10580,9452,3648,2793,7467,4276,681,9043,4445,863,631,683,3380,1420,803,12720,421,741,5610,6397,1741,4478,3090,2167,5872,2575,2138,9380,4672,743,1919,552,1056,700,436,1041,7199,1085,596,636,988,3921,4619,696,1114,844,773,764,7059,896,1007,593,561,4971,11188,422,372,2423,4819,5152,5359,9738,1659,17513,5565,766,4939,855,545,612,2745,9541,854,802,13646,6904,1435,8639,684,16650,814,15035,944,1614,1435,625,2699,828,6686,7443,663,3759,5706,8983,2258,4830,2085,2233,3195,9523,1766,2234,905,9009,1694,7246,18648,435,2442,7805,872,1254,2800,1238,836,15246,13320,1949,12549,625,4749,2259,1107,1410,7841,12734,2839,658,2629,974,4473,876,18255,3783,6989,3295,576,596,394,2102,1053,1304,710,1343,1365,1443,5550,975,1008,459,6301,1594,4520,781,711,1812,620,805,447,1153,830,710,1450,10739,705,3502,417,13420,1244,646,5381,421,679,6344,4309,1641,863,2017,808,1748,7999,489,6962,673,14543,12707,8569,4861,574,1656,1657,1289,4355,1323,15188,576,4446,15189,669,8896,5294,6618,2255,2484,449,2880,1986,3167,15654,686,3024,4355,441,7162,942,3323,4791,1046,552,4432,10038,5364,2551,491,2047,5556,670,3135,894,1407,13564,5698,705,945,863,803,813,789,830,3738,1055,2161,993,6629,645,10851,3095,10053,2988,2347,3378,2190,16062,789,673,12171,1562,1792,507,362,4788,1370,6517,4062,1154,600,16778,3271,1837,827,8334,967,10662,3966,8184,4210,6103,17209,1968,8501,641,880,7903,13293,5058,746,1864,1021,2761,2011,816,2452,855,613,1846,658,702,932,1892,702,6453,6013,1024,6762,783,683,526,2569,1124,668,827,16872,813,2599,610,2594,1140,1580,470,432,4137,12338,670,1993,787,973,3485,5701,4562,2288,13027,7812,700,1760,4427,4576,1381,815,5980,2518,4480,3856,3475,803,2404,872,530,4131,1624,2219,10134,6019,10738,1571,6996,12196,2479,810,6169,2393,702,2656,5653,1015,608,5799,9710,1956,7334,6019,10636,5497,1882,2415,2807,4282,816,794,821,918,895,6473,408,4026,4601,17579,1024,2269,401,1223,1686,618,651,664,1327,6132,8260,730,3499,746,1080,863,1132,2206,12644,3895,1814,5606,608,3427,544,612,5953,1853,942,7451,716,827,722,15510,1741,7978,802,2310,993,464,3104,1221,2295,548,2810,391,5646,1419,1001,620,2587,5487,648,1207,761,9574,14674,3394,5269,675,571,18067,727,3967,862,842,854,9471,505,1752,10283,7006,12738,5967,4282,1665,2484,873,789,4795,7963,756,658,2616,956,1136,851,3064,1983,1103,2002,4162,734,789,14027,2981,10462,965,3528,7094,1322,4243,2312,9537,5058,648,17073,506,4718,827,2776,13669,791,5932,8341,6960,4032,2444,6793,1871,5174,4466,7207,462,5288,2016,4129,1079,2106,3923,2960,6534,10562,9474,3437,9089,1169,1326,7774,1085,847,14294,1190,3755,3192,1800,2564,2042,5946,3387,1821,1715,698,1586,890,2960,612,653,1206,852,1624,14968,3026,1625,5698,1607,18440,15714,1610,900,1056,3535,2674,755,1974,990,4478,5151,6963,3405,4933,1020,526,1700,6271,2481,1637,2977,1664,10888,449,1240,880,789,1111,4749,7548,827,523,7553,997,11796,571,802,6094,17849,470,450,1273,2309,608,2711,1065,2331,13500,900,3484,5247,2382,705,13182,1744,1985,5526,7534,939,830,1401,4368,608,1781,537,680,6088,852,928,864,1814,12459,548,13499,13254,3096,802,4517,5600,729,8836,6814,3941,5799,857,4844,5430,3948,3946,5437,1384,868,625,7603,1399,1211,5688,936,7143,3671,499,1250,576,4762,1610,4688,694,3761,5938,2680,9555,6406,2446,435,956,480,9556,1569,2805,7003,450,11189,776,7173,1963,603,864,3864,4973,1830,757,14833,2945,13248,4181,5973,10947,776,473,17590,675,7010,583,464,593,648,1161,3053,3290,701,968,3544,2585,5264,3570,1721,2396,3506,9576,405,9768,13661,2939,921,837,828,3345,568,4341,582,790,665,7518,7466,3959,6663,4662,2394,1410,8975,3925,4315,4200,2328,642,5171,1621,1043,4586,1148,11106,2285,3124,1564,816,706,958,1910,1698,13043,3024,1235,797,2112,1153,4690,3249,608,596,788,2578,3445,15218,15688,2058,853,1953,4899,13111,8280,728,761,5125,1244,853,5021,1177,2234,6288,3745,972,614,837,658,14066,4466,624,2564,8743,1623,4315,16111,717,9158,2466,851,1836,13325,487,4006,4480,7251,12648,4472,5125,6467,4257,698,15466,1822,2762,11688,7676,5002,593,652,720,645,4109,1577,645,1082,4725,459,2387,945,8121,4729,972,3136,1387,4688,900,665,652,2863,4221,1362,9533,1196,1381,3816,5533,9218,4612,473,7273,6066,4879,1190,740,684,2275,683,8261,7128,2511,407,4596,7290,596,2040,1238,1607,2437,906,11192,4879,449,2442,6743,2045,465,5174,4926,6701,9203,12968,720,666,511,3849,4183,2505,1083,1902,852,2215,2354,1033,1026,2642,680,5351,9704,3660,15105,1644,5218,18176,1003,3385,7673,11871,3043,1752,5140,2184,11765,1893,579,969,2489,1228,4844,1569,620,6160,6239,1656,523,1017,3553,17455,876,765,17569,6966,526,1443,11233,3455,4851,827,15851,2821,457,1715,963,1662,954,3176,1760,2583,3190,1048,10935,1303,10937,4596,6407,3667,4476,787,579,454,4989,1024,5291,4830,1031,1289,878,1000,7372,773,3278,2681,15451,1279,478,587,4381,5569,521,17522,1207,2406,988,1662,5067,725,1031,1172,4639,2211,1356,2562,2617,16599,776,7139,4890,777,5697,4088,16737,3006,2647,362,13720,2368,4981,17724,3175,2095,12907,811,5455,689,7966,3071,984,1565,4939,8476,4328,3888,4418,743,432,2369,8930,749,516,17634,10204,4851,1595,6073,3878,2753,935,521,9281,7136,3673,1997,2096,5167,10313,16169,719,8736,2889,584,3293,4319,1838,2228,761,6296,1624,4772,548,18426,5492,1961,5898,645,1893,2832,1151,8128,6573,3160,921,5017,495,4937,1439,868,4702,1289,5167,972,4665,7325,3105,2115,2184,1914,1121,1167,10032,6036,12554,1116,12581,1154,9705,3013,7400,3740,805,454,4320,4108,644,3407,9904,3872,11382,2104,1351,1412,394,4566,1628,11544,3150,3350,7942,775,6042,12141,6335,5334,16626,654,622,4095,3226,14027,721,770,10012,5242,12831,779,5950,7305,1059,10539,2644,844,577,2148,432,666,1991,942,2686,4633,4195,2883,2710,6199,2999,485,1389,3701,2897,1097,3570,4471,8013,1882,3796,5016,3595,12499,5408,947,11584,695,620,8127,4004,485,2923,2913,982,434,7415,932,18741,492,12430,5252,3455,9581,537,743,2196,2159,1624,804,844,1389,17825,4633,6335,9513,16261,1740,6702,1063,1132,801,2489,1160,12121,2692,2207,863,3352,3468,693,18395,10401,3626,1031,5100,3685,2973,18756,6112,2475,434,1167,2258,8994,4724,1427,4514,758,6743,421,628,675,975,9081,9734,1949,9960,14593,3264,664,1200,2175,7706,8183,3360,5893,1842,6925,4755,4150,532,900,12219,697,10907,3049,3806,2200,1963,3620,10886,772,1801,4620,1026,5699,2428,3447,5142,5889,1363,2647,5132,759,11549,844,7333,8698,8505,675,463,2386,1009,1438,4965,751,951,4232,5940,8752,828,2863,1187,1881,1626,1020,2808,847,764,3162,3655,11954,3435,8540,771,12677,12923,6039,1637,1615,4486,793,1591,419,4033,3080,766,772,9510,741,571,970,7220,2426,11214,6031,4562,1989,802,2064,4869,891,752,491,12071,13161,998,2744,1048,3802,2717,748,820,1187,7677,4864,1059,969,2100,1007,3280,651,4740,12832,9609,2841,4821,3913,552,2329,1115,3478,4447,7931,842,8213,1166,2777,16779,432,653,963,11644,895,9569,1624,11147,7730,2356,7079,15824,1307,6751,3871,4742,4311,788,3136,2239,775,1208,6425,2699,1657,4504,1050,2117,1140,8674,702,477,658,875,9513,668,2428,841,5042,2359,11146,2467,893,1279,1401,1139,612,2394,1401,5006,17889,737,5220,1343,1264,8930,6108,6968,734,850,526,5863,631,1687,4044,1966,1442,557,5593,4704,2425,603,17688,3418,830,1111,507,4582,6662,518,983,945,956,1271,1436,4395,942,2482,2271,789,10424,4315,4883,2913,492,679,1975,492,6648,7466,4032,5817,2064,579,2873,614,1637,1115,7032,6479,1107,1757,544,14386,506,6751,4047,942,561,1809,17029,4584,4229,918,3215,2157,5505,1370,780,1875,7476,5229,1914,984,1026,1436,487,1727,4191,2319,799,2496,1738,767,1728,9408,6682,9058,4412,485,518,3763,490,10090,3172,2217,8233,3606,949,2827,6798,3669,8931,1103,526,469,3749,1827,5914,2923,3808,2387,5717,336,1875,2400,658,5941,574,7550,11235,5010,4611,8227,391,1079,1454,8774,530,945,944,6296,14603,990,1172,2112,2822,4186,4754,1088,2699,3810,597,4253,6857,708,12747,5912,1128,7167,4807,12677,1878,4816,1073,2912,4435,6588,755,4318,3381,17425,1689,4854,936,2609,731,447,1004,1921,6776,2212,5805,952,2227,4440,1114,1286,18429,8982,6288,4180,1717,706,4233,5319,5002,1569,14542,789,625,8032,5241,755,5433,761,1392,561,9720,1166,777,579,1373,776,1316,5296,2120,11088,1807,13098,2150,3266,2789,3176,918,2801,2795,1194,1092,756,16309,902,7453,2642,3197,743,3453,8996,4284,898,5292,2912,18055,2331,7875,1180,734,827,4712,645,2079,3924,7098,801,669,7955,4242,838,507,713,6416,4158,1153,2319,5134,4584,18398,745,3220,1821,4858,635,816,800,689,4847,1257,1179,8820,1827,1215,2407,1599,15247,393,5504,488,6777,789,438,5480,926,15231,6871,1219,1749,863,1118,861,1075,1165,1605,3496,5044,1035,1046,6539,2591,763,2270,784,734,17235,1333,745,9122,9013,7797,7752,8366,6480,5189,491,868,4816,539,6797,1240,3118,5557,872,730,8257,2371,6330,7260,652,1274,1052,12338,6195,2324,2382,17216,6737,1815,3353,4508,418,828,5219,2657,4697,2688,900,11119,3010,9961,545,5792,1394,617,4290,3694,593,1235,3103,1845,2301,4030,687,3107,2658,666,645,7076,5363,1436,5346,668,540,2596,5163,612,530,506,5443,3920,1786,7247,614,2866,461,842,1918,1778,11392,3663,720,17955,2939,470,1777,6020,4039,2131,942,912,13994,936,15197,1832,2551,1046,2095,1002,3478,2511,789,9796,1179,2176,639,11764,2239,1114,5439,5897,1662,720,8316,489,5312,18293,5825,17084,5841,1299,1624,863,11525,3077,16314,6140,1949,16353,13631,1914,5301,1175,756,5119,906,10771,2148,2760,904,16582,6091,9697,5880,864,926,5606,1232,5739,2016,678,2066,4565,569,524,4101,2680,523,17849,3603,7870,2314,914,8114,2399,5005,9377,432,2414,687,1857,912,521,9533,4284,734,4230,9419,9641,422,2972,5183,17294,18419,14745,2753,765,7662,1649,898,17882,4939,3078,11946,4295,4077,6966,540,7408,17650,589,4222,2074,1239,530,2261,1569,1334,838,15968,17153,737,645,5378,1743,1588,6781,7655,505,2078,4252,1367,2570,1057,1274,834,5065,628,1131,4155,553,898,10184,2719,3299,7811,5038,1237,17279,15511,1235,10274,2426,589,4771,4653,7466,1815,13512,608,12210,394,7844,421,12958,1198,5422,5743,1600,3226,3979,2445,6989,1229,1089,1237,754,4844,2079,700,3787,7258,10993,4558,1872,1059,6500,6140,4200,1613,4668,11586,4679,906,774,4001,2601,1431,1682,789,7131,730,1947,448,552,384,5718,550,10577,5308,2865,8110,4507,898,10824,2037,6499,15324,767,915,738,4578,6295,5198,3774,1592,596,9289,477,11078,16957,933,593,2765,756,1754,3799,17987,2933,1637,900,5067,373,1002,867,1600,2306,781,3061,904,3557,4588,8627,4946,1814,4173,3802,2130,1698,882,5233,504,679,2717,1031,10115,6588,14201,2919,1030,1723,795,3018,8492,7610,1923,13872,12182,7611,1393,5458,1257,687,2583,4749,2814,1822,12047,7162,776,1000,1050,746,473,984,1257,6285,15467,3256,2184,8529,2443,2278,8113,633,4530,1052,1551,3462,4191,7942,11279,1662,663,6313,1873,785,948,7056,394,4789,3502,470,7918,745,2610,3917,12236,1655,2365,462,10742,3462,4562,2443,561,1050,965,3035,10217,4466,2607,1308,447,11554,847,1353,852,4774,1098,3160,1606,2834,14138,878,1671,7308,7679,850,7589,1621,526,4191,12725,2392,6982,4155,10512,6669,3599,15272,1014,6564,2329,2549,1018,2400,5518,1334,663,590,2841,4537,6438,984,1177,848,605,1668,600,2066,5002,4196,2556,8167,4916,1644,7600,921,2711,868,814,1046,6188,2016,2345,1694,810,2467,394,3884,945,2088,14024,8275,2529,3890,18440,1745,5884,3811,754,2209,995,449,1550,3601,3304,2527,16277,3342,10091,5140,4468,2676,4563,3198,877,684,3183,484,435,931,4743,1133,702,1053,3801,1935,15065,608,2815,13052,743,690,833,4162,873,12610,1666,3042,579,2469,689,17203,14386,13753,4791,7351,6470,3941,5421,1173,2352,743,459,1408,5759,15843,1179,872,4183,706,2104,662,886,3965,9461,5672,1142,10389,8151,800,2647,487,4189,1002,1026,17534,463,1333,4564,5544,2877,531,12621,1236,1041,2442,722,9572,1155,851,2901,3884,4712,5599,8808,521,6303,2815,2781,473,988,842,1291,660,5759,936,11957,2623,7637,10023,5804,6662,1205,3012,470,1867,507,1613,505,2705,1040,2137,1004,4538,2805,2153,4385,2801,9278,3437,793,6825,7589,2557,9552,1656,4096,15984,3284,2469,4383,2826,3562,802,765,2838,950,574,4439,9245,4779,521,5592,1243,6196,1715,1262,1270,4554,505,540,5707,3037,5838,3075,6458,5819,13037,4078,13813,6980,773,1750,1628,3618,480,3871,515,1817,5171,6062,4113,1060,8281,776,1421,2295,2819,737,13502,1129,726,4719,3063,4013,2035,6411,1323,9694,647,3991,1227,1689,633,2018,1676,2039,984,4627,2958,1691,4435,600,2269,786,2646,544,949,2724,11096,2372,9877,984,4853,2657,1882,4092,4498,3624,2036,1847,778,16041,11382,777,1848,5330,8138,3133,3996,1018,12981,1168,3370,5031,945,943,2441,1247,3922,2218,4081,8303,2082,921,561,10738,7929,11105,889,3360,984,9525,6509,1184,2517,669,3006,477,532,544,1254,1250,682,868,9526,10915,8119,1808,743,8220,5556,1810,4277,10388,3091,14792,4223,2447,5639,1881,2386,4682,10226,4497,949,4129,15095,544,3730,505,833,828,3265,3121,564,702,936,5164,4374,710,878,2795,2451,2230,2293,612,4666,3567,7258,1958,3035,2329,15955,5038,14803,2005,551,684,1024,952,4149,1266,990,1676,419,4203,625,868,602,1935,7848,7577,6928,1838,2877,1829,481,3057,1445,620,3965,13903,1662,3791,16874,1875,5154,1071,1374,946,3247,758,10051,658,6516,11434,10337,1797,8349,4751,949,755,3781,1727,517,2237,4077,4241,1356,4679,2344,4688,4619,552,2579,790,6857,1916,766,2428,2517,515,3775,2313,12787,1433,477,886,713,563,12862,14236,5387,1122,689,1264,3731,5042,8067,2095,780,9281,3595,506,14488,8596,2360,596,3312,707,12874,925,2400,2481,1172,5267,4980,9952,1577,1781,9797,700,992,3506,3889,990,2927,708,938,4323,1030,12947,684,10309,633,4560,645,8521,4140,684,805,13406,5285,907,729,925,534,1698,716,612,1192,5241,13912,767,7715,7238,1819,1624,1957,707,730,9439,4503,1808,1281,633,645,1915,928,798,1245,2624,5484,4687,5557,1355,4826,9680,723,1332,5969,692,4838,545,411,7480,911,538,10577,433,7127,1129,921,2718,3605,573,743,3618,1051,12267,4124,4123,6887,485,15303,5012,702,4921,6132,3462,8964,4514,1782,9896,2751,1888,2301,3763,16538,891,5412,427,457,2494,6125,707,470,6156,12799,915,1415,11301,1079,1046,9140,1799,4504,649,711,14482,6694,3854,6012,839,622,624,6377,4724,574,4082,5645,3350,923,5816,4268,9230,7014,819,18363,1588,5169,3376,2287,743,2298,766,1415,2401,1612,931,626,1410,1061,983,4090,579,1715,524,3210,4353,794,3332,18172,12437,828,1238,4398,1115,9780,1013,13099,581,4140,2300,12848,4321,6442,17175,5670,2009,949,7677,11530,1424,1884,3546,7081,16492,787,4127,18236,870,10093,1632,1358,610,842,3590,6337,1809,4484,3105,15134,1680,3545,2015,8014,5107,8216,2345,816,678,5450,1076,1085,1013,13588,957,3656,6377,3489,5087,12320,817,17747,2427,603,9541,13034,886,1046,10428,688,462,912,8314,7455],"mode":"markers","hoverinfo":["text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text","text"],"text":["Carat: 0.73 <br /> Price: 2397 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 3300 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 713 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 707 <br /> Clarity: VVS1","Carat: 0.31 <br /> Price: 987 <br /> Clarity: IF","Carat: 0.83 <br /> Price: 3250 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 1668 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 1771 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 1053 <br /> Clarity: VS1","Carat: 1.1 <br /> Price: 4640 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 2467 <br /> Clarity: VVS2","Carat: 0.32 <br /> Price: 758 <br /> Clarity: VS2","Carat: 1 <br /> Price: 5755 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 6097 <br /> Clarity: VS2","Carat: 2.01 <br /> Price: 17422 <br /> Clarity: VS2","Carat: 1.6 <br /> Price: 12343 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4525 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 734 <br /> Clarity: VS2","Carat: 0.38 <br /> Price: 685 <br /> Clarity: VS2","Carat: 1.03 <br /> Price: 4326 <br /> Clarity: SI1","Carat: 1.31 <br /> Price: 5546 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2100 <br /> Clarity: SI1","Carat: 2.01 <br /> Price: 12829 <br /> Clarity: SI2","Carat: 0.8 <br /> Price: 4002 <br /> Clarity: VVS2","Carat: 1.2 <br /> Price: 7274 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 698 <br /> Clarity: SI1","Carat: 0.82 <br /> Price: 2643 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 683 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 1207 <br /> Clarity: IF","Carat: 0.33 <br /> Price: 403 <br /> Clarity: SI2","Carat: 1.3 <br /> Price: 5824 <br /> Clarity: VS2","Carat: 0.35 <br /> Price: 1116 <br /> Clarity: IF","Carat: 1.25 <br /> Price: 4200 <br /> Clarity: SI1","Carat: 0.62 <br /> Price: 1441 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 545 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 673 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 5013 <br /> Clarity: VS1","Carat: 0.4 <br /> Price: 1351 <br /> Clarity: VVS2","Carat: 0.5 <br /> Price: 1374 <br /> Clarity: SI1","Carat: 1.09 <br /> Price: 5219 <br /> Clarity: SI1","Carat: 0.91 <br /> Price: 3985 <br /> Clarity: SI2","Carat: 0.34 <br /> Price: 829 <br /> Clarity: VS2","Carat: 1.66 <br /> Price: 9882 <br /> Clarity: VVS2","Carat: 0.42 <br /> Price: 1103 <br /> Clarity: VS1","Carat: 1.17 <br /> Price: 4639 <br /> Clarity: SI2","Carat: 1.27 <br /> Price: 8312 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 4579 <br /> Clarity: VS1","Carat: 1.55 <br /> Price: 16137 <br /> Clarity: VS2","Carat: 1.5 <br /> Price: 9173 <br /> Clarity: SI2","Carat: 1.06 <br /> Price: 5817 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 3950 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 1656 <br /> Clarity: VVS1","Carat: 0.32 <br /> Price: 534 <br /> Clarity: VS2","Carat: 0.38 <br /> Price: 1400 <br /> Clarity: VVS1","Carat: 0.78 <br /> Price: 2386 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 682 <br /> Clarity: VVS1","Carat: 2.01 <br /> Price: 16733 <br /> Clarity: SI2","Carat: 0.34 <br /> Price: 596 <br /> Clarity: VS2","Carat: 1.31 <br /> Price: 6089 <br /> Clarity: SI2","Carat: 0.9 <br /> Price: 3748 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 532 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 802 <br /> Clarity: VS1","Carat: 1.22 <br /> Price: 6456 <br /> Clarity: SI1","Carat: 2.15 <br /> Price: 14388 <br /> Clarity: SI2","Carat: 1.27 <br /> Price: 5761 <br /> Clarity: VVS1","Carat: 1.06 <br /> Price: 5783 <br /> Clarity: VS2","Carat: 2 <br /> Price: 11200 <br /> Clarity: SI1","Carat: 2.01 <br /> Price: 12845 <br /> Clarity: VS1","Carat: 0.23 <br /> Price: 472 <br /> Clarity: VVS1","Carat: 1.16 <br /> Price: 5852 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 6975 <br /> Clarity: VVS2","Carat: 0.34 <br /> Price: 596 <br /> Clarity: SI1","Carat: 0.55 <br /> Price: 2042 <br /> Clarity: VVS1","Carat: 0.43 <br /> Price: 1143 <br /> Clarity: VVS1","Carat: 0.5 <br /> Price: 875 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 1013 <br /> Clarity: VVS1","Carat: 1.52 <br /> Price: 8818 <br /> Clarity: SI1","Carat: 1.22 <br /> Price: 6969 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 766 <br /> Clarity: VVS2","Carat: 0.5 <br /> Price: 965 <br /> Clarity: VS2","Carat: 1.26 <br /> Price: 9243 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 1153 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 734 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1395 <br /> Clarity: VS1","Carat: 0.39 <br /> Price: 988 <br /> Clarity: VVS2","Carat: 0.31 <br /> Price: 840 <br /> Clarity: VVS1","Carat: 1.3 <br /> Price: 7747 <br /> Clarity: VS1","Carat: 1.05 <br /> Price: 4974 <br /> Clarity: SI1","Carat: 1.06 <br /> Price: 6619 <br /> Clarity: VS2","Carat: 0.51 <br /> Price: 1569 <br /> Clarity: VS2","Carat: 0.52 <br /> Price: 1721 <br /> Clarity: VS1","Carat: 1.52 <br /> Price: 16670 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 523 <br /> Clarity: SI2","Carat: 1.33 <br /> Price: 15031 <br /> Clarity: VVS2","Carat: 0.41 <br /> Price: 953 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 662 <br /> Clarity: VVS2","Carat: 0.88 <br /> Price: 3472 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 645 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2548 <br /> Clarity: SI1","Carat: 0.53 <br /> Price: 1412 <br /> Clarity: VS2","Carat: 0.52 <br /> Price: 1232 <br /> Clarity: VS2","Carat: 0.69 <br /> Price: 1863 <br /> Clarity: VS2","Carat: 0.65 <br /> Price: 1881 <br /> Clarity: VS2","Carat: 1.2 <br /> Price: 5868 <br /> Clarity: SI1","Carat: 1 <br /> Price: 7056 <br /> Clarity: VS1","Carat: 1.2 <br /> Price: 2376 <br /> Clarity: I1","Carat: 1.5 <br /> Price: 16116 <br /> Clarity: VVS2","Carat: 0.38 <br /> Price: 1000 <br /> Clarity: VS1","Carat: 1.51 <br /> Price: 10781 <br /> Clarity: SI2","Carat: 0.35 <br /> Price: 460 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 555 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4853 <br /> Clarity: SI1","Carat: 0.54 <br /> Price: 1944 <br /> Clarity: VVS1","Carat: 0.31 <br /> Price: 734 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1323 <br /> Clarity: SI1","Carat: 1.04 <br /> Price: 4508 <br /> Clarity: SI2","Carat: 1.19 <br /> Price: 5937 <br /> Clarity: SI1","Carat: 2.13 <br /> Price: 15110 <br /> Clarity: VS2","Carat: 1.1 <br /> Price: 5958 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 514 <br /> Clarity: SI1","Carat: 1 <br /> Price: 4081 <br /> Clarity: SI2","Carat: 0.71 <br /> Price: 2686 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 4113 <br /> Clarity: SI2","Carat: 0.51 <br /> Price: 2501 <br /> Clarity: VVS1","Carat: 0.71 <br /> Price: 2839 <br /> Clarity: VS1","Carat: 0.55 <br /> Price: 1901 <br /> Clarity: VS1","Carat: 0.34 <br /> Price: 765 <br /> Clarity: SI1","Carat: 0.79 <br /> Price: 2827 <br /> Clarity: VVS2","Carat: 0.36 <br /> Price: 789 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 731 <br /> Clarity: VS1","Carat: 0.89 <br /> Price: 3429 <br /> Clarity: SI2","Carat: 0.57 <br /> Price: 2546 <br /> Clarity: VS2","Carat: 1.22 <br /> Price: 6250 <br /> Clarity: SI2","Carat: 1.09 <br /> Price: 8057 <br /> Clarity: VVS2","Carat: 0.54 <br /> Price: 1745 <br /> Clarity: VS1","Carat: 1.02 <br /> Price: 3142 <br /> Clarity: SI2","Carat: 0.9 <br /> Price: 3160 <br /> Clarity: SI2","Carat: 1.14 <br /> Price: 7667 <br /> Clarity: VS2","Carat: 0.26 <br /> Price: 453 <br /> Clarity: VS1","Carat: 0.54 <br /> Price: 1268 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 4296 <br /> Clarity: VS2","Carat: 1 <br /> Price: 5938 <br /> Clarity: VS2","Carat: 2.02 <br /> Price: 12485 <br /> Clarity: SI2","Carat: 1.5 <br /> Price: 14199 <br /> Clarity: VS2","Carat: 0.53 <br /> Price: 1173 <br /> Clarity: VS2","Carat: 1 <br /> Price: 4844 <br /> Clarity: SI1","Carat: 2.3 <br /> Price: 18108 <br /> Clarity: SI2","Carat: 1 <br /> Price: 5292 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 8637 <br /> Clarity: SI1","Carat: 0.62 <br /> Price: 2367 <br /> Clarity: IF","Carat: 0.81 <br /> Price: 2926 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 768 <br /> Clarity: VS1","Carat: 2 <br /> Price: 10494 <br /> Clarity: I1","Carat: 0.31 <br /> Price: 710 <br /> Clarity: VVS1","Carat: 1.01 <br /> Price: 5599 <br /> Clarity: SI1","Carat: 0.57 <br /> Price: 2172 <br /> Clarity: VVS1","Carat: 0.77 <br /> Price: 2257 <br /> Clarity: SI1","Carat: 1.03 <br /> Price: 7533 <br /> Clarity: VS2","Carat: 1.23 <br /> Price: 5622 <br /> Clarity: VS2","Carat: 1.24 <br /> Price: 8073 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 2381 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 607 <br /> Clarity: VS1","Carat: 1.04 <br /> Price: 4084 <br /> Clarity: SI2","Carat: 1.19 <br /> Price: 4508 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 999 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 702 <br /> Clarity: SI1","Carat: 1 <br /> Price: 3819 <br /> Clarity: SI2","Carat: 0.53 <br /> Price: 1607 <br /> Clarity: VS2","Carat: 0.27 <br /> Price: 459 <br /> Clarity: VVS2","Carat: 1.11 <br /> Price: 5281 <br /> Clarity: SI1","Carat: 1.63 <br /> Price: 9256 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 3061 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 1289 <br /> Clarity: IF","Carat: 1.73 <br /> Price: 9271 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 1187 <br /> Clarity: IF","Carat: 1.3 <br /> Price: 14196 <br /> Clarity: VVS1","Carat: 0.24 <br /> Price: 478 <br /> Clarity: VVS1","Carat: 0.35 <br /> Price: 1116 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 776 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4816 <br /> Clarity: SI1","Carat: 0.27 <br /> Price: 383 <br /> Clarity: SI1","Carat: 1.16 <br /> Price: 4872 <br /> Clarity: SI1","Carat: 0.59 <br /> Price: 1771 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2751 <br /> Clarity: VS1","Carat: 0.5 <br /> Price: 1559 <br /> Clarity: VS2","Carat: 1.5 <br /> Price: 10962 <br /> Clarity: SI1","Carat: 0.55 <br /> Price: 3468 <br /> Clarity: VVS2","Carat: 0.36 <br /> Price: 538 <br /> Clarity: SI1","Carat: 1.05 <br /> Price: 4363 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 675 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 4871 <br /> Clarity: SI1","Carat: 1 <br /> Price: 7145 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 1662 <br /> Clarity: VS2","Carat: 0.81 <br /> Price: 2616 <br /> Clarity: SI2","Carat: 0.72 <br /> Price: 2125 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 692 <br /> Clarity: VVS2","Carat: 0.23 <br /> Price: 402 <br /> Clarity: VS1","Carat: 0.51 <br /> Price: 1749 <br /> Clarity: VS1","Carat: 0.84 <br /> Price: 2656 <br /> Clarity: SI2","Carat: 0.59 <br /> Price: 1643 <br /> Clarity: VS2","Carat: 0.91 <br /> Price: 4256 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 1591 <br /> Clarity: VS2","Carat: 0.87 <br /> Price: 3664 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 526 <br /> Clarity: SI1","Carat: 0.96 <br /> Price: 3355 <br /> Clarity: SI1","Carat: 0.63 <br /> Price: 2391 <br /> Clarity: VS2","Carat: 1.09 <br /> Price: 12185 <br /> Clarity: VVS2","Carat: 0.92 <br /> Price: 3787 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 5056 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 814 <br /> Clarity: VVS2","Carat: 0.31 <br /> Price: 628 <br /> Clarity: VS2","Carat: 1 <br /> Price: 3819 <br /> Clarity: VS2","Carat: 2.2 <br /> Price: 15092 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4969 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 2266 <br /> Clarity: VS1","Carat: 0.72 <br /> Price: 2670 <br /> Clarity: SI2","Carat: 1.31 <br /> Price: 12008 <br /> Clarity: VS1","Carat: 0.77 <br /> Price: 3189 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 1599 <br /> Clarity: VS1","Carat: 0.59 <br /> Price: 3026 <br /> Clarity: VVS2","Carat: 1.54 <br /> Price: 8408 <br /> Clarity: SI2","Carat: 0.57 <br /> Price: 1642 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 2686 <br /> Clarity: VS2","Carat: 0.51 <br /> Price: 1974 <br /> Clarity: VVS2","Carat: 1.31 <br /> Price: 7277 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 3959 <br /> Clarity: SI1","Carat: 1 <br /> Price: 7350 <br /> Clarity: VS1","Carat: 1.5 <br /> Price: 13853 <br /> Clarity: VS2","Carat: 1.06 <br /> Price: 5516 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 1030 <br /> Clarity: VVS2","Carat: 0.34 <br /> Price: 875 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 558 <br /> Clarity: VS2","Carat: 1.11 <br /> Price: 5450 <br /> Clarity: VS2","Carat: 0.41 <br /> Price: 1356 <br /> Clarity: VVS1","Carat: 0.72 <br /> Price: 2642 <br /> Clarity: VVS1","Carat: 0.32 <br /> Price: 648 <br /> Clarity: VS1","Carat: 0.53 <br /> Price: 1775 <br /> Clarity: VS1","Carat: 0.74 <br /> Price: 2936 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 783 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 2648 <br /> Clarity: VS1","Carat: 1.5 <br /> Price: 12265 <br /> Clarity: SI1","Carat: 1.13 <br /> Price: 4689 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1436 <br /> Clarity: VS2","Carat: 0.41 <br /> Price: 1015 <br /> Clarity: SI1","Carat: 1.04 <br /> Price: 3512 <br /> Clarity: SI2","Carat: 0.92 <br /> Price: 4309 <br /> Clarity: SI2","Carat: 1.5 <br /> Price: 10147 <br /> Clarity: IF","Carat: 0.9 <br /> Price: 3114 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 523 <br /> Clarity: VS1","Carat: 1.14 <br /> Price: 9090 <br /> Clarity: VS1","Carat: 0.58 <br /> Price: 2528 <br /> Clarity: IF","Carat: 1.57 <br /> Price: 12809 <br /> Clarity: VS2","Carat: 0.91 <br /> Price: 4031 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2353 <br /> Clarity: VVS2","Carat: 0.7 <br /> Price: 2113 <br /> Clarity: SI1","Carat: 0.92 <br /> Price: 3282 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 638 <br /> Clarity: VVS2","Carat: 2 <br /> Price: 11975 <br /> Clarity: SI2","Carat: 1.32 <br /> Price: 8501 <br /> Clarity: VVS1","Carat: 0.58 <br /> Price: 1903 <br /> Clarity: VS2","Carat: 1.05 <br /> Price: 5821 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2092 <br /> Clarity: SI2","Carat: 0.35 <br /> Price: 647 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2500 <br /> Clarity: VVS2","Carat: 1.5 <br /> Price: 13853 <br /> Clarity: VS2","Carat: 0.58 <br /> Price: 1184 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 3869 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 493 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 972 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 3448 <br /> Clarity: VS2","Carat: 1.52 <br /> Price: 11021 <br /> Clarity: VVS2","Carat: 0.7 <br /> Price: 2576 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 1792 <br /> Clarity: SI2","Carat: 0.92 <br /> Price: 3724 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 4760 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 596 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 854 <br /> Clarity: VS1","Carat: 1.5 <br /> Price: 7560 <br /> Clarity: SI2","Carat: 0.42 <br /> Price: 1031 <br /> Clarity: VS1","Carat: 1.05 <br /> Price: 5468 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 698 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 1811 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 926 <br /> Clarity: VVS2","Carat: 1 <br /> Price: 4956 <br /> Clarity: SI2","Carat: 1.02 <br /> Price: 6652 <br /> Clarity: VS2","Carat: 0.27 <br /> Price: 393 <br /> Clarity: SI1","Carat: 1.08 <br /> Price: 4544 <br /> Clarity: SI2","Carat: 0.43 <br /> Price: 1016 <br /> Clarity: SI1","Carat: 1.13 <br /> Price: 4025 <br /> Clarity: SI2","Carat: 1.5 <br /> Price: 11360 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 612 <br /> Clarity: VS1","Carat: 0.56 <br /> Price: 1224 <br /> Clarity: VS2","Carat: 1.2 <br /> Price: 13015 <br /> Clarity: VVS1","Carat: 0.36 <br /> Price: 663 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 854 <br /> Clarity: VVS1","Carat: 0.51 <br /> Price: 1899 <br /> Clarity: IF","Carat: 2.07 <br /> Price: 13786 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 2788 <br /> Clarity: I1","Carat: 0.33 <br /> Price: 579 <br /> Clarity: VS2","Carat: 0.6 <br /> Price: 1820 <br /> Clarity: VS1","Carat: 0.6 <br /> Price: 2130 <br /> Clarity: VVS2","Carat: 0.35 <br /> Price: 1116 <br /> Clarity: IF","Carat: 0.75 <br /> Price: 2992 <br /> Clarity: VVS2","Carat: 1.12 <br /> Price: 8973 <br /> Clarity: IF","Carat: 0.26 <br /> Price: 554 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 4653 <br /> Clarity: SI1","Carat: 0.43 <br /> Price: 1062 <br /> Clarity: VVS2","Carat: 0.27 <br /> Price: 470 <br /> Clarity: VS2","Carat: 1 <br /> Price: 3634 <br /> Clarity: SI2","Carat: 1.03 <br /> Price: 6539 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 3676 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 781 <br /> Clarity: VS2","Carat: 0.24 <br /> Price: 419 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 6204 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 368 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 4594 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 17153 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1354 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 552 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 2590 <br /> Clarity: VS2","Carat: 1.11 <br /> Price: 6563 <br /> Clarity: SI1","Carat: 0.28 <br /> Price: 429 <br /> Clarity: VS1","Carat: 1.29 <br /> Price: 2596 <br /> Clarity: I1","Carat: 0.57 <br /> Price: 1359 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 723 <br /> Clarity: VS2","Carat: 0.75 <br /> Price: 2940 <br /> Clarity: VS2","Carat: 0.35 <br /> Price: 906 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 781 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 645 <br /> Clarity: VS2","Carat: 0.29 <br /> Price: 619 <br /> Clarity: VS1","Carat: 1.03 <br /> Price: 6786 <br /> Clarity: VS2","Carat: 1.7 <br /> Price: 12117 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 3312 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 758 <br /> Clarity: VS1","Carat: 0.61 <br /> Price: 1438 <br /> Clarity: VS2","Carat: 0.51 <br /> Price: 1687 <br /> Clarity: SI1","Carat: 1.13 <br /> Price: 4535 <br /> Clarity: SI2","Carat: 1.02 <br /> Price: 4381 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 956 <br /> Clarity: IF","Carat: 0.31 <br /> Price: 625 <br /> Clarity: VVS2","Carat: 1.51 <br /> Price: 3734 <br /> Clarity: I1","Carat: 0.81 <br /> Price: 3402 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 1935 <br /> Clarity: SI2","Carat: 0.74 <br /> Price: 2282 <br /> Clarity: SI2","Carat: 0.51 <br /> Price: 1841 <br /> Clarity: VS1","Carat: 0.35 <br /> Price: 984 <br /> Clarity: VS2","Carat: 0.62 <br /> Price: 1378 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 576 <br /> Clarity: SI2","Carat: 0.58 <br /> Price: 1953 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4306 <br /> Clarity: SI1","Carat: 1.07 <br /> Price: 6885 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 2838 <br /> Clarity: SI1","Carat: 1.04 <br /> Price: 4427 <br /> Clarity: SI2","Carat: 0.51 <br /> Price: 1879 <br /> Clarity: IF","Carat: 0.53 <br /> Price: 1813 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 675 <br /> Clarity: VS1","Carat: 0.9 <br /> Price: 4309 <br /> Clarity: VS1","Carat: 0.78 <br /> Price: 3253 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 738 <br /> Clarity: VS2","Carat: 1.51 <br /> Price: 7145 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 4238 <br /> Clarity: SI2","Carat: 0.43 <br /> Price: 919 <br /> Clarity: SI1","Carat: 1 <br /> Price: 4939 <br /> Clarity: SI1","Carat: 0.38 <br /> Price: 871 <br /> Clarity: VS1","Carat: 0.6 <br /> Price: 1562 <br /> Clarity: SI2","Carat: 0.54 <br /> Price: 1669 <br /> Clarity: IF","Carat: 0.49 <br /> Price: 1225 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 4657 <br /> Clarity: VVS2","Carat: 1.23 <br /> Price: 8145 <br /> Clarity: VS1","Carat: 1.21 <br /> Price: 5893 <br /> Clarity: VVS1","Carat: 0.7 <br /> Price: 2135 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 914 <br /> Clarity: IF","Carat: 0.56 <br /> Price: 2157 <br /> Clarity: IF","Carat: 1.71 <br /> Price: 13445 <br /> Clarity: VS2","Carat: 0.25 <br /> Price: 548 <br /> Clarity: VVS2","Carat: 0.35 <br /> Price: 906 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 1018 <br /> Clarity: IF","Carat: 0.4 <br /> Price: 828 <br /> Clarity: IF","Carat: 0.82 <br /> Price: 3674 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 3382 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 999 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 878 <br /> Clarity: VVS1","Carat: 1.5 <br /> Price: 9157 <br /> Clarity: SI2","Carat: 0.76 <br /> Price: 3249 <br /> Clarity: VS2","Carat: 0.51 <br /> Price: 1627 <br /> Clarity: VS1","Carat: 0.4 <br /> Price: 938 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 2375 <br /> Clarity: SI1","Carat: 0.24 <br /> Price: 419 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 1060 <br /> Clarity: IF","Carat: 0.31 <br /> Price: 593 <br /> Clarity: SI1","Carat: 0.73 <br /> Price: 3002 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 784 <br /> Clarity: VS2","Carat: 0.28 <br /> Price: 646 <br /> Clarity: VVS2","Carat: 1.17 <br /> Price: 4826 <br /> Clarity: SI2","Carat: 1.5 <br /> Price: 10428 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 8475 <br /> Clarity: SI1","Carat: 1.26 <br /> Price: 4742 <br /> Clarity: SI2","Carat: 1.18 <br /> Price: 3278 <br /> Clarity: I1","Carat: 1.02 <br /> Price: 3519 <br /> Clarity: I1","Carat: 0.26 <br /> Price: 657 <br /> Clarity: VVS1","Carat: 0.4 <br /> Price: 702 <br /> Clarity: SI1","Carat: 1 <br /> Price: 4543 <br /> Clarity: SI1","Carat: 0.35 <br /> Price: 979 <br /> Clarity: IF","Carat: 1.01 <br /> Price: 4269 <br /> Clarity: SI2","Carat: 0.34 <br /> Price: 760 <br /> Clarity: VS1","Carat: 0.73 <br /> Price: 3307 <br /> Clarity: VS2","Carat: 1 <br /> Price: 7114 <br /> Clarity: VS2","Carat: 0.45 <br /> Price: 1282 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 2817 <br /> Clarity: SI1","Carat: 2.12 <br /> Price: 18118 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 526 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 1193 <br /> Clarity: IF","Carat: 0.32 <br /> Price: 540 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 418 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 668 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 5919 <br /> Clarity: VS2","Carat: 1.52 <br /> Price: 15649 <br /> Clarity: VS1","Carat: 1.77 <br /> Price: 14561 <br /> Clarity: SI1","Carat: 1.08 <br /> Price: 4173 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 1052 <br /> Clarity: IF","Carat: 0.32 <br /> Price: 758 <br /> Clarity: VS2","Carat: 0.27 <br /> Price: 622 <br /> Clarity: VVS2","Carat: 0.32 <br /> Price: 684 <br /> Clarity: SI1","Carat: 1.24 <br /> Price: 10395 <br /> Clarity: VVS2","Carat: 0.36 <br /> Price: 821 <br /> Clarity: VVS2","Carat: 0.71 <br /> Price: 2291 <br /> Clarity: SI2","Carat: 0.9 <br /> Price: 2732 <br /> Clarity: VS1","Carat: 1.47 <br /> Price: 7438 <br /> Clarity: SI2","Carat: 0.56 <br /> Price: 2442 <br /> Clarity: VVS2","Carat: 1.8 <br /> Price: 14959 <br /> Clarity: VS1","Carat: 2.02 <br /> Price: 9095 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 710 <br /> Clarity: VS2","Carat: 0.38 <br /> Price: 941 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 907 <br /> Clarity: VVS1","Carat: 0.91 <br /> Price: 4429 <br /> Clarity: SI1","Carat: 0.39 <br /> Price: 919 <br /> Clarity: SI1","Carat: 0.49 <br /> Price: 949 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 698 <br /> Clarity: VS2","Carat: 0.55 <br /> Price: 1833 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 854 <br /> Clarity: VS1","Carat: 0.91 <br /> Price: 2700 <br /> Clarity: SI1","Carat: 0.73 <br /> Price: 2871 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 898 <br /> Clarity: SI1","Carat: 1.3 <br /> Price: 7333 <br /> Clarity: VS1","Carat: 1.24 <br /> Price: 6383 <br /> Clarity: VVS2","Carat: 0.72 <br /> Price: 1814 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 4989 <br /> Clarity: SI1","Carat: 1.72 <br /> Price: 12451 <br /> Clarity: VS1","Carat: 0.72 <br /> Price: 1799 <br /> Clarity: SI2","Carat: 1.21 <br /> Price: 4492 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 957 <br /> Clarity: IF","Carat: 0.71 <br /> Price: 2516 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 1206 <br /> Clarity: I1","Carat: 0.73 <br /> Price: 3089 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 11322 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 4972 <br /> Clarity: SI1","Carat: 2 <br /> Price: 17247 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 7182 <br /> Clarity: SI2","Carat: 0.71 <br /> Price: 3082 <br /> Clarity: VS1","Carat: 0.34 <br /> Price: 765 <br /> Clarity: VVS2","Carat: 1.29 <br /> Price: 3774 <br /> Clarity: I1","Carat: 1 <br /> Price: 4416 <br /> Clarity: SI1","Carat: 0.72 <br /> Price: 2139 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 499 <br /> Clarity: SI1","Carat: 1.17 <br /> Price: 5423 <br /> Clarity: SI2","Carat: 0.5 <br /> Price: 1883 <br /> Clarity: VVS1","Carat: 0.7 <br /> Price: 2020 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 873 <br /> Clarity: IF","Carat: 0.31 <br /> Price: 507 <br /> Clarity: VS2","Carat: 2.04 <br /> Price: 14527 <br /> Clarity: VS1","Carat: 1.33 <br /> Price: 6405 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 480 <br /> Clarity: SI2","Carat: 1 <br /> Price: 5292 <br /> Clarity: SI1","Carat: 1.52 <br /> Price: 8868 <br /> Clarity: SI2","Carat: 0.71 <br /> Price: 3465 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 976 <br /> Clarity: VVS1","Carat: 0.33 <br /> Price: 965 <br /> Clarity: VVS2","Carat: 0.34 <br /> Price: 974 <br /> Clarity: IF","Carat: 0.34 <br /> Price: 540 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 7741 <br /> Clarity: VVS1","Carat: 1.52 <br /> Price: 6793 <br /> Clarity: I1","Carat: 2.02 <br /> Price: 16256 <br /> Clarity: SI2","Carat: 1.64 <br /> Price: 16297 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 1200 <br /> Clarity: VS1","Carat: 1 <br /> Price: 4528 <br /> Clarity: SI1","Carat: 0.38 <br /> Price: 1069 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 638 <br /> Clarity: VVS2","Carat: 1.1 <br /> Price: 5610 <br /> Clarity: SI1","Carat: 1.08 <br /> Price: 5559 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 6333 <br /> Clarity: VS2","Carat: 0.84 <br /> Price: 3685 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 7695 <br /> Clarity: SI2","Carat: 2.01 <br /> Price: 15841 <br /> Clarity: SI2","Carat: 0.71 <br /> Price: 2686 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 645 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 393 <br /> Clarity: VS2","Carat: 1.19 <br /> Price: 5622 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4480 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 2398 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 489 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 941 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2229 <br /> Clarity: SI2","Carat: 1.25 <br /> Price: 5331 <br /> Clarity: VS1","Carat: 0.87 <br /> Price: 4189 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 13853 <br /> Clarity: VS2","Carat: 0.59 <br /> Price: 1821 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4766 <br /> Clarity: SI1","Carat: 0.36 <br /> Price: 905 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 462 <br /> Clarity: SI2","Carat: 0.78 <br /> Price: 2652 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2536 <br /> Clarity: VVS2","Carat: 0.39 <br /> Price: 855 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 3231 <br /> Clarity: VS1","Carat: 1.07 <br /> Price: 5967 <br /> Clarity: VS1","Carat: 0.4 <br /> Price: 877 <br /> Clarity: VS1","Carat: 1.04 <br /> Price: 7375 <br /> Clarity: VS1","Carat: 0.36 <br /> Price: 1035 <br /> Clarity: VVS1","Carat: 0.81 <br /> Price: 3266 <br /> Clarity: VS1","Carat: 0.52 <br /> Price: 1743 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 625 <br /> Clarity: VVS2","Carat: 1.1 <br /> Price: 9257 <br /> Clarity: VVS1","Carat: 0.41 <br /> Price: 683 <br /> Clarity: SI2","Carat: 1.07 <br /> Price: 7440 <br /> Clarity: VS1","Carat: 0.73 <br /> Price: 3250 <br /> Clarity: VS2","Carat: 0.23 <br /> Price: 530 <br /> Clarity: VVS2","Carat: 0.32 <br /> Price: 828 <br /> Clarity: VS2","Carat: 0.26 <br /> Price: 554 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 608 <br /> Clarity: VS1","Carat: 0.33 <br /> Price: 1002 <br /> Clarity: VVS2","Carat: 0.4 <br /> Price: 1125 <br /> Clarity: VS1","Carat: 1 <br /> Price: 4630 <br /> Clarity: VS1","Carat: 0.73 <br /> Price: 3589 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 1869 <br /> Clarity: VS2","Carat: 0.38 <br /> Price: 969 <br /> Clarity: SI1","Carat: 0.39 <br /> Price: 936 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 1172 <br /> Clarity: VVS2","Carat: 0.35 <br /> Price: 644 <br /> Clarity: SI1","Carat: 0.55 <br /> Price: 2129 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 7306 <br /> Clarity: VVS2","Carat: 0.42 <br /> Price: 921 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 840 <br /> Clarity: VVS1","Carat: 0.43 <br /> Price: 867 <br /> Clarity: VS1","Carat: 1.12 <br /> Price: 8430 <br /> Clarity: VVS2","Carat: 0.51 <br /> Price: 1928 <br /> Clarity: VS2","Carat: 0.52 <br /> Price: 978 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 776 <br /> Clarity: VVS2","Carat: 0.44 <br /> Price: 941 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4498 <br /> Clarity: SI2","Carat: 0.9 <br /> Price: 4068 <br /> Clarity: SI1","Carat: 1.56 <br /> Price: 12146 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 6221 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 579 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 610 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 3145 <br /> Clarity: VS2","Carat: 1.7 <br /> Price: 10165 <br /> Clarity: VS1","Carat: 2.02 <br /> Price: 11390 <br /> Clarity: SI2","Carat: 2.17 <br /> Price: 13782 <br /> Clarity: SI2","Carat: 0.28 <br /> Price: 787 <br /> Clarity: IF","Carat: 0.53 <br /> Price: 1268 <br /> Clarity: SI1","Carat: 1.59 <br /> Price: 9209 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1140 <br /> Clarity: SI1","Carat: 1.07 <br /> Price: 4949 <br /> Clarity: SI2","Carat: 0.53 <br /> Price: 2051 <br /> Clarity: VVS2","Carat: 0.42 <br /> Price: 884 <br /> Clarity: VVS1","Carat: 0.33 <br /> Price: 713 <br /> Clarity: VVS1","Carat: 1.08 <br /> Price: 6532 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 505 <br /> Clarity: VS2","Carat: 0.44 <br /> Price: 1063 <br /> Clarity: VVS2","Carat: 0.73 <br /> Price: 2801 <br /> Clarity: VS2","Carat: 0.84 <br /> Price: 3015 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 755 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 779 <br /> Clarity: VVS2","Carat: 0.31 <br /> Price: 593 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 675 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 8214 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1024 <br /> Clarity: SI2","Carat: 1.04 <br /> Price: 5070 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 596 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 1122 <br /> Clarity: IF","Carat: 0.41 <br /> Price: 1115 <br /> Clarity: VVS1","Carat: 1.51 <br /> Price: 11255 <br /> Clarity: SI1","Carat: 0.91 <br /> Price: 3734 <br /> Clarity: VS1","Carat: 1.22 <br /> Price: 5226 <br /> Clarity: SI2","Carat: 1.2 <br /> Price: 6167 <br /> Clarity: VS1","Carat: 0.51 <br /> Price: 1591 <br /> Clarity: VS2","Carat: 1.51 <br /> Price: 13068 <br /> Clarity: VS2","Carat: 0.81 <br /> Price: 2534 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 734 <br /> Clarity: VS1","Carat: 1.24 <br /> Price: 6232 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 6075 <br /> Clarity: SI1","Carat: 0.36 <br /> Price: 801 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2145 <br /> Clarity: VS1","Carat: 1.04 <br /> Price: 4811 <br /> Clarity: SI2","Carat: 0.5 <br /> Price: 1909 <br /> Clarity: VVS2","Carat: 1.02 <br /> Price: 8818 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 628 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 1011 <br /> Clarity: VVS1","Carat: 1.12 <br /> Price: 4064 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 675 <br /> Clarity: SI1","Carat: 0.27 <br /> Price: 623 <br /> Clarity: VVS2","Carat: 0.68 <br /> Price: 2516 <br /> Clarity: VS1","Carat: 2.14 <br /> Price: 13540 <br /> Clarity: SI2","Carat: 0.53 <br /> Price: 844 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2753 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 802 <br /> Clarity: VVS2","Carat: 0.7 <br /> Price: 2106 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 686 <br /> Clarity: VS2","Carat: 0.41 <br /> Price: 613 <br /> Clarity: VS2","Carat: 0.57 <br /> Price: 1949 <br /> Clarity: VS2","Carat: 1 <br /> Price: 4265 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 548 <br /> Clarity: VS1","Carat: 0.74 <br /> Price: 1865 <br /> Clarity: I1","Carat: 0.9 <br /> Price: 4286 <br /> Clarity: SI1","Carat: 0.53 <br /> Price: 2051 <br /> Clarity: VVS2","Carat: 1.3 <br /> Price: 5242 <br /> Clarity: VS2","Carat: 1 <br /> Price: 4514 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 727 <br /> Clarity: I1","Carat: 0.95 <br /> Price: 3590 <br /> Clarity: SI2","Carat: 2.01 <br /> Price: 15116 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 1870 <br /> Clarity: VS1","Carat: 1.11 <br /> Price: 4212 <br /> Clarity: SI2","Carat: 0.75 <br /> Price: 3336 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 18159 <br /> Clarity: VVS2","Carat: 0.72 <br /> Price: 2857 <br /> Clarity: SI1","Carat: 0.74 <br /> Price: 2222 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2636 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 5443 <br /> Clarity: SI1","Carat: 0.24 <br /> Price: 678 <br /> Clarity: VVS1","Carat: 1.01 <br /> Price: 6075 <br /> Clarity: SI1","Carat: 0.43 <br /> Price: 943 <br /> Clarity: VS1","Carat: 0.43 <br /> Price: 669 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 1061 <br /> Clarity: VS2","Carat: 1.24 <br /> Price: 5889 <br /> Clarity: SI2","Carat: 2.03 <br /> Price: 18257 <br /> Clarity: SI1","Carat: 2.01 <br /> Price: 13387 <br /> Clarity: SI1","Carat: 1.23 <br /> Price: 5269 <br /> Clarity: SI2","Carat: 0.78 <br /> Price: 2874 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 667 <br /> Clarity: VS2","Carat: 0.63 <br /> Price: 2879 <br /> Clarity: IF","Carat: 2.32 <br /> Price: 18026 <br /> Clarity: SI1","Carat: 1.12 <br /> Price: 5818 <br /> Clarity: SI1","Carat: 0.56 <br /> Price: 1686 <br /> Clarity: VS1","Carat: 1.59 <br /> Price: 17366 <br /> Clarity: VS1","Carat: 1.04 <br /> Price: 6542 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 6104 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 756 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2290 <br /> Clarity: VS1","Carat: 0.42 <br /> Price: 933 <br /> Clarity: VVS2","Carat: 1.5 <br /> Price: 7368 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 656 <br /> Clarity: VS2","Carat: 1 <br /> Price: 3906 <br /> Clarity: SI2","Carat: 2.04 <br /> Price: 15092 <br /> Clarity: SI2","Carat: 1.05 <br /> Price: 9092 <br /> Clarity: VVS2","Carat: 1.53 <br /> Price: 7617 <br /> Clarity: SI2","Carat: 1.48 <br /> Price: 8815 <br /> Clarity: SI1","Carat: 1.12 <br /> Price: 5845 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4899 <br /> Clarity: SI1","Carat: 0.82 <br /> Price: 3050 <br /> Clarity: SI2","Carat: 1.51 <br /> Price: 11512 <br /> Clarity: VS1","Carat: 0.56 <br /> Price: 2040 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 670 <br /> Clarity: VS1","Carat: 0.25 <br /> Price: 459 <br /> Clarity: VS2","Carat: 0.93 <br /> Price: 3839 <br /> Clarity: SI2","Carat: 1.08 <br /> Price: 4124 <br /> Clarity: SI2","Carat: 0.6 <br /> Price: 1196 <br /> Clarity: SI2","Carat: 1.53 <br /> Price: 14483 <br /> Clarity: VS2","Carat: 1.5 <br /> Price: 10886 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 1050 <br /> Clarity: VVS2","Carat: 0.9 <br /> Price: 3734 <br /> Clarity: IF","Carat: 1.03 <br /> Price: 5063 <br /> Clarity: SI1","Carat: 1.23 <br /> Price: 7988 <br /> Clarity: VS2","Carat: 0.84 <br /> Price: 4365 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 3304 <br /> Clarity: VS2","Carat: 0.26 <br /> Price: 584 <br /> Clarity: VVS2","Carat: 0.37 <br /> Price: 741 <br /> Clarity: VS1","Carat: 0.52 <br /> Price: 1911 <br /> Clarity: VVS2","Carat: 2.11 <br /> Price: 14740 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 10688 <br /> Clarity: SI1","Carat: 1 <br /> Price: 6047 <br /> Clarity: VS2","Carat: 0.55 <br /> Price: 3005 <br /> Clarity: VVS1","Carat: 0.51 <br /> Price: 2208 <br /> Clarity: IF","Carat: 1.01 <br /> Price: 5204 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 814 <br /> Clarity: VVS1","Carat: 0.58 <br /> Price: 1359 <br /> Clarity: SI2","Carat: 1.26 <br /> Price: 6733 <br /> Clarity: SI1","Carat: 0.78 <br /> Price: 2486 <br /> Clarity: SI2","Carat: 0.84 <br /> Price: 4068 <br /> Clarity: SI1","Carat: 0.23 <br /> Price: 530 <br /> Clarity: VVS1","Carat: 1.5 <br /> Price: 13603 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 854 <br /> Clarity: VS1","Carat: 0.51 <br /> Price: 2369 <br /> Clarity: VVS1","Carat: 0.73 <br /> Price: 3184 <br /> Clarity: VS2","Carat: 0.52 <br /> Price: 1273 <br /> Clarity: SI2","Carat: 2.04 <br /> Price: 15618 <br /> Clarity: SI2","Carat: 1.54 <br /> Price: 14433 <br /> Clarity: VS1","Carat: 1.53 <br /> Price: 7677 <br /> Clarity: VS1","Carat: 0.24 <br /> Price: 478 <br /> Clarity: VVS2","Carat: 0.55 <br /> Price: 2030 <br /> Clarity: VS2","Carat: 0.52 <br /> Price: 1372 <br /> Clarity: SI2","Carat: 0.44 <br /> Price: 1235 <br /> Clarity: VVS1","Carat: 1.19 <br /> Price: 7633 <br /> Clarity: VS2","Carat: 0.35 <br /> Price: 898 <br /> Clarity: IF","Carat: 1.76 <br /> Price: 9659 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 3167 <br /> Clarity: I1","Carat: 0.41 <br /> Price: 1061 <br /> Clarity: VS2","Carat: 0.76 <br /> Price: 3833 <br /> Clarity: VVS2","Carat: 0.5 <br /> Price: 1348 <br /> Clarity: VS2","Carat: 1.09 <br /> Price: 5219 <br /> Clarity: SI1","Carat: 1.21 <br /> Price: 3862 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 1815 <br /> Clarity: VS2","Carat: 0.39 <br /> Price: 1053 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1128 <br /> Clarity: SI1","Carat: 0.92 <br /> Price: 3555 <br /> Clarity: SI2","Carat: 0.37 <br /> Price: 833 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2673 <br /> Clarity: VS2","Carat: 1.71 <br /> Price: 13068 <br /> Clarity: VS2","Carat: 1.21 <br /> Price: 8879 <br /> Clarity: VS1","Carat: 0.52 <br /> Price: 2338 <br /> Clarity: VVS2","Carat: 0.51 <br /> Price: 1365 <br /> Clarity: VS2","Carat: 1.2 <br /> Price: 5376 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 698 <br /> Clarity: VVS2","Carat: 0.41 <br /> Price: 1076 <br /> Clarity: VVS2","Carat: 0.81 <br /> Price: 2604 <br /> Clarity: SI2","Carat: 0.72 <br /> Price: 2804 <br /> Clarity: SI1","Carat: 0.83 <br /> Price: 2811 <br /> Clarity: SI1","Carat: 0.63 <br /> Price: 2357 <br /> Clarity: VS2","Carat: 1.8 <br /> Price: 12338 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 1170 <br /> Clarity: IF","Carat: 0.34 <br /> Price: 803 <br /> Clarity: SI1","Carat: 2.14 <br /> Price: 15769 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2932 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 796 <br /> Clarity: IF","Carat: 1.21 <br /> Price: 10706 <br /> Clarity: VS1","Carat: 0.54 <br /> Price: 1259 <br /> Clarity: VS2","Carat: 0.41 <br /> Price: 1076 <br /> Clarity: VVS2","Carat: 0.43 <br /> Price: 716 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 421 <br /> Clarity: SI2","Carat: 0.24 <br /> Price: 449 <br /> Clarity: VVS2","Carat: 1 <br /> Price: 7190 <br /> Clarity: VS1","Carat: 0.51 <br /> Price: 1121 <br /> Clarity: SI2","Carat: 0.8 <br /> Price: 3653 <br /> Clarity: VS2","Carat: 1.54 <br /> Price: 12308 <br /> Clarity: IF","Carat: 0.74 <br /> Price: 3163 <br /> Clarity: VS2","Carat: 1.23 <br /> Price: 9836 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 828 <br /> Clarity: VS1","Carat: 0.29 <br /> Price: 511 <br /> Clarity: VVS2","Carat: 1 <br /> Price: 3360 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 1015 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 847 <br /> Clarity: VVS1","Carat: 0.55 <br /> Price: 1062 <br /> Clarity: VS2","Carat: 0.57 <br /> Price: 1655 <br /> Clarity: VS2","Carat: 0.39 <br /> Price: 734 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 701 <br /> Clarity: SI1","Carat: 0.82 <br /> Price: 2799 <br /> Clarity: SI2","Carat: 1.5 <br /> Price: 16409 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 2264 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 805 <br /> Clarity: VS2","Carat: 1.03 <br /> Price: 5581 <br /> Clarity: SI1","Carat: 1.03 <br /> Price: 8972 <br /> Clarity: VVS2","Carat: 1.04 <br /> Price: 8921 <br /> Clarity: IF","Carat: 0.61 <br /> Price: 2331 <br /> Clarity: VVS2","Carat: 0.31 <br /> Price: 766 <br /> Clarity: VVS2","Carat: 0.72 <br /> Price: 2314 <br /> Clarity: SI1","Carat: 0.98 <br /> Price: 2898 <br /> Clarity: SI2","Carat: 0.29 <br /> Price: 607 <br /> Clarity: VVS2","Carat: 0.27 <br /> Price: 622 <br /> Clarity: VVS1","Carat: 1 <br /> Price: 3672 <br /> Clarity: SI2","Carat: 0.38 <br /> Price: 1008 <br /> Clarity: VVS2","Carat: 1.83 <br /> Price: 10043 <br /> Clarity: VS2","Carat: 0.51 <br /> Price: 1662 <br /> Clarity: VS2","Carat: 1.16 <br /> Price: 5908 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1554 <br /> Clarity: VS2","Carat: 1.52 <br /> Price: 12418 <br /> Clarity: SI1","Carat: 0.91 <br /> Price: 4513 <br /> Clarity: SI1","Carat: 1.04 <br /> Price: 10804 <br /> Clarity: VVS2","Carat: 1.02 <br /> Price: 4583 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 6429 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2371 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 642 <br /> Clarity: VS1","Carat: 0.4 <br /> Price: 1078 <br /> Clarity: VVS2","Carat: 0.65 <br /> Price: 1945 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 945 <br /> Clarity: I1","Carat: 0.3 <br /> Price: 886 <br /> Clarity: IF","Carat: 1.02 <br /> Price: 7257 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 926 <br /> Clarity: VVS1","Carat: 0.71 <br /> Price: 1883 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 492 <br /> Clarity: SI1","Carat: 0.38 <br /> Price: 967 <br /> Clarity: VVS1","Carat: 0.51 <br /> Price: 1202 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 574 <br /> Clarity: SI1","Carat: 0.54 <br /> Price: 1637 <br /> Clarity: SI1","Carat: 1 <br /> Price: 4623 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 776 <br /> Clarity: VVS2","Carat: 0.31 <br /> Price: 802 <br /> Clarity: VS2","Carat: 1.51 <br /> Price: 13162 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 3713 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 435 <br /> Clarity: VS2","Carat: 0.76 <br /> Price: 3192 <br /> Clarity: VS2","Carat: 1.12 <br /> Price: 4811 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 2573 <br /> Clarity: SI1","Carat: 0.55 <br /> Price: 2112 <br /> Clarity: VS2","Carat: 1.12 <br /> Price: 7781 <br /> Clarity: VS1","Carat: 0.58 <br /> Price: 1442 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 1367 <br /> Clarity: IF","Carat: 1.02 <br /> Price: 6738 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 2198 <br /> Clarity: SI1","Carat: 1.05 <br /> Price: 7070 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 752 <br /> Clarity: VVS2","Carat: 0.7 <br /> Price: 2307 <br /> Clarity: SI2","Carat: 1 <br /> Price: 4732 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 608 <br /> Clarity: VS1","Carat: 1 <br /> Price: 5671 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 11577 <br /> Clarity: VS2","Carat: 0.86 <br /> Price: 3284 <br /> Clarity: SI2","Carat: 0.26 <br /> Price: 482 <br /> Clarity: VS1","Carat: 0.35 <br /> Price: 906 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1316 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 9253 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 5226 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 720 <br /> Clarity: VS1","Carat: 1 <br /> Price: 5522 <br /> Clarity: SI1","Carat: 0.39 <br /> Price: 588 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 705 <br /> Clarity: SI1","Carat: 1.2 <br /> Price: 6578 <br /> Clarity: SI1","Carat: 1 <br /> Price: 4872 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4751 <br /> Clarity: SI2","Carat: 0.73 <br /> Price: 2093 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 698 <br /> Clarity: VS2","Carat: 1 <br /> Price: 4602 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 838 <br /> Clarity: IF","Carat: 0.58 <br /> Price: 1132 <br /> Clarity: SI1","Carat: 0.93 <br /> Price: 3437 <br /> Clarity: SI1","Carat: 1.31 <br /> Price: 5869 <br /> Clarity: SI1","Carat: 1.04 <br /> Price: 7049 <br /> Clarity: VS1","Carat: 0.42 <br /> Price: 1103 <br /> Clarity: VVS2","Carat: 1.04 <br /> Price: 5265 <br /> Clarity: SI2","Carat: 1.02 <br /> Price: 6352 <br /> Clarity: VS2","Carat: 0.79 <br /> Price: 2784 <br /> Clarity: VS1","Carat: 0.38 <br /> Price: 680 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 5372 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 6563 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 1002 <br /> Clarity: VS2","Carat: 0.67 <br /> Price: 1897 <br /> Clarity: SI1","Carat: 0.44 <br /> Price: 1003 <br /> Clarity: VS2","Carat: 1.23 <br /> Price: 5894 <br /> Clarity: SI2","Carat: 0.52 <br /> Price: 1956 <br /> Clarity: IF","Carat: 0.7 <br /> Price: 2026 <br /> Clarity: SI2","Carat: 0.8 <br /> Price: 5007 <br /> Clarity: VS1","Carat: 0.33 <br /> Price: 540 <br /> Clarity: SI1","Carat: 1 <br /> Price: 5420 <br /> Clarity: VS2","Carat: 0.37 <br /> Price: 763 <br /> Clarity: VS2","Carat: 0.74 <br /> Price: 2715 <br /> Clarity: VVS1","Carat: 1.2 <br /> Price: 4838 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 583 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 2368 <br /> Clarity: VS2","Carat: 0.51 <br /> Price: 1552 <br /> Clarity: VS2","Carat: 0.76 <br /> Price: 2847 <br /> Clarity: SI1","Carat: 1.27 <br /> Price: 7125 <br /> Clarity: SI1","Carat: 1.7 <br /> Price: 15662 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 544 <br /> Clarity: VS1","Carat: 1.53 <br /> Price: 14931 <br /> Clarity: VS1","Carat: 0.83 <br /> Price: 3347 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 1694 <br /> Clarity: VS2","Carat: 1.06 <br /> Price: 4167 <br /> Clarity: SI2","Carat: 0.53 <br /> Price: 1273 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 8951 <br /> Clarity: SI2","Carat: 1.51 <br /> Price: 8313 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 3045 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 537 <br /> Clarity: VS1","Carat: 1.52 <br /> Price: 17659 <br /> Clarity: VS1","Carat: 0.9 <br /> Price: 3613 <br /> Clarity: SI2","Carat: 1.54 <br /> Price: 11177 <br /> Clarity: VS2","Carat: 0.56 <br /> Price: 1824 <br /> Clarity: VS1","Carat: 1 <br /> Price: 5505 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 7487 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 790 <br /> Clarity: VVS2","Carat: 1.2 <br /> Price: 5818 <br /> Clarity: VS1","Carat: 0.75 <br /> Price: 3011 <br /> Clarity: SI1","Carat: 0.62 <br /> Price: 1410 <br /> Clarity: SI2","Carat: 1.16 <br /> Price: 5793 <br /> Clarity: SI2","Carat: 0.82 <br /> Price: 3012 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 862 <br /> Clarity: VVS1","Carat: 1.52 <br /> Price: 13215 <br /> Clarity: VS2","Carat: 0.91 <br /> Price: 4670 <br /> Clarity: VS1","Carat: 0.37 <br /> Price: 957 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 842 <br /> Clarity: VS1","Carat: 1.11 <br /> Price: 5102 <br /> Clarity: SI1","Carat: 1 <br /> Price: 3360 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4064 <br /> Clarity: SI2","Carat: 1.5 <br /> Price: 14092 <br /> Clarity: VS2","Carat: 0.58 <br /> Price: 1758 <br /> Clarity: SI1","Carat: 1.06 <br /> Price: 4815 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 8371 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 645 <br /> Clarity: VS1","Carat: 0.91 <br /> Price: 4357 <br /> Clarity: VS2","Carat: 0.51 <br /> Price: 1744 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 433 <br /> Clarity: SI2","Carat: 0.38 <br /> Price: 700 <br /> Clarity: VS2","Carat: 0.27 <br /> Price: 432 <br /> Clarity: VS1","Carat: 0.9 <br /> Price: 3368 <br /> Clarity: SI2","Carat: 0.78 <br /> Price: 2431 <br /> Clarity: SI2","Carat: 0.55 <br /> Price: 1402 <br /> Clarity: VS2","Carat: 0.44 <br /> Price: 733 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 3063 <br /> Clarity: VS1","Carat: 0.54 <br /> Price: 2041 <br /> Clarity: VS1","Carat: 1.04 <br /> Price: 2037 <br /> Clarity: I1","Carat: 1.04 <br /> Price: 4973 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2348 <br /> Clarity: SI1","Carat: 0.43 <br /> Price: 1045 <br /> Clarity: SI1","Carat: 1.04 <br /> Price: 4403 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 529 <br /> Clarity: VS1","Carat: 0.8 <br /> Price: 2935 <br /> Clarity: VS1","Carat: 0.38 <br /> Price: 1067 <br /> Clarity: VS1","Carat: 0.5 <br /> Price: 1760 <br /> Clarity: VVS2","Carat: 1.51 <br /> Price: 7553 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1554 <br /> Clarity: VS2","Carat: 1.19 <br /> Price: 4168 <br /> Clarity: SI2","Carat: 0.94 <br /> Price: 4106 <br /> Clarity: SI1","Carat: 0.45 <br /> Price: 1263 <br /> Clarity: VVS1","Carat: 0.32 <br /> Price: 421 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 1229 <br /> Clarity: IF","Carat: 0.25 <br /> Price: 407 <br /> Clarity: VS2","Carat: 0.79 <br /> Price: 2976 <br /> Clarity: SI1","Carat: 1.52 <br /> Price: 9642 <br /> Clarity: SI1","Carat: 1.22 <br /> Price: 5160 <br /> Clarity: VS2","Carat: 0.43 <br /> Price: 1318 <br /> Clarity: IF","Carat: 0.31 <br /> Price: 734 <br /> Clarity: VS2","Carat: 1.08 <br /> Price: 6426 <br /> Clarity: SI1","Carat: 0.43 <br /> Price: 1358 <br /> Clarity: VVS1","Carat: 0.7 <br /> Price: 2348 <br /> Clarity: SI2","Carat: 1.21 <br /> Price: 7806 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 540 <br /> Clarity: SI1","Carat: 1.22 <br /> Price: 6201 <br /> Clarity: VS1","Carat: 1.21 <br /> Price: 10351 <br /> Clarity: VS1","Carat: 1 <br /> Price: 4379 <br /> Clarity: SI2","Carat: 0.74 <br /> Price: 2956 <br /> Clarity: VS1","Carat: 1.5 <br /> Price: 6770 <br /> Clarity: SI2","Carat: 1.64 <br /> Price: 8794 <br /> Clarity: SI2","Carat: 0.93 <br /> Price: 3101 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 707 <br /> Clarity: VVS2","Carat: 0.91 <br /> Price: 3363 <br /> Clarity: SI1","Carat: 0.37 <br /> Price: 811 <br /> Clarity: VS2","Carat: 1.21 <br /> Price: 4946 <br /> Clarity: SI2","Carat: 0.73 <br /> Price: 4196 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 911 <br /> Clarity: VS2","Carat: 2.02 <br /> Price: 18565 <br /> Clarity: VS2","Carat: 1.21 <br /> Price: 6261 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 1158 <br /> Clarity: IF","Carat: 1.06 <br /> Price: 5250 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 394 <br /> Clarity: SI2","Carat: 1.57 <br /> Price: 9478 <br /> Clarity: VS2","Carat: 0.72 <br /> Price: 2802 <br /> Clarity: VS2","Carat: 0.39 <br /> Price: 1248 <br /> Clarity: IF","Carat: 0.41 <br /> Price: 1419 <br /> Clarity: IF","Carat: 0.38 <br /> Price: 1030 <br /> Clarity: VVS2","Carat: 0.31 <br /> Price: 872 <br /> Clarity: VS2","Carat: 0.65 <br /> Price: 1708 <br /> Clarity: SI1","Carat: 1.32 <br /> Price: 6079 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 3775 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 936 <br /> Clarity: VVS1","Carat: 1.2 <br /> Price: 5638 <br /> Clarity: SI1","Carat: 0.46 <br /> Price: 1816 <br /> Clarity: VVS1","Carat: 0.77 <br /> Price: 3697 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4749 <br /> Clarity: SI1","Carat: 1.12 <br /> Price: 3700 <br /> Clarity: SI2","Carat: 1.56 <br /> Price: 12082 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 731 <br /> Clarity: VS1","Carat: 0.72 <br /> Price: 1907 <br /> Clarity: SI2","Carat: 1.02 <br /> Price: 3601 <br /> Clarity: SI2","Carat: 1 <br /> Price: 6245 <br /> Clarity: VS2","Carat: 2.22 <br /> Price: 18706 <br /> Clarity: VS1","Carat: 0.53 <br /> Price: 1803 <br /> Clarity: VS2","Carat: 1.22 <br /> Price: 6887 <br /> Clarity: SI1","Carat: 0.56 <br /> Price: 1427 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 984 <br /> Clarity: IF","Carat: 2.24 <br /> Price: 16709 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 743 <br /> Clarity: VS2","Carat: 1.05 <br /> Price: 6486 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 734 <br /> Clarity: VS1","Carat: 0.73 <br /> Price: 2826 <br /> Clarity: VS1","Carat: 2.09 <br /> Price: 13092 <br /> Clarity: SI2","Carat: 1.09 <br /> Price: 4538 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2453 <br /> Clarity: SI2","Carat: 0.57 <br /> Price: 1785 <br /> Clarity: VS2","Carat: 0.73 <br /> Price: 2846 <br /> Clarity: VVS2","Carat: 0.33 <br /> Price: 743 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 911 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 492 <br /> Clarity: SI2","Carat: 1.73 <br /> Price: 8816 <br /> Clarity: SI2","Carat: 0.51 <br /> Price: 1383 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 732 <br /> Clarity: SI1","Carat: 0.57 <br /> Price: 1573 <br /> Clarity: VS2","Carat: 0.8 <br /> Price: 2862 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 2735 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 672 <br /> Clarity: VS2","Carat: 1.25 <br /> Price: 10285 <br /> Clarity: VS1","Carat: 0.34 <br /> Price: 641 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 796 <br /> Clarity: VVS1","Carat: 0.68 <br /> Price: 2074 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 14465 <br /> Clarity: VS2","Carat: 0.52 <br /> Price: 2012 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 878 <br /> Clarity: VVS2","Carat: 0.52 <br /> Price: 1577 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 548 <br /> Clarity: VS2","Carat: 1.23 <br /> Price: 6403 <br /> Clarity: SI2","Carat: 1 <br /> Price: 3465 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 3431 <br /> Clarity: VS1","Carat: 0.33 <br /> Price: 1010 <br /> Clarity: IF","Carat: 1.58 <br /> Price: 10345 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2290 <br /> Clarity: VS2","Carat: 1.27 <br /> Price: 6654 <br /> Clarity: SI1","Carat: 0.37 <br /> Price: 703 <br /> Clarity: VS1","Carat: 0.56 <br /> Price: 1091 <br /> Clarity: SI2","Carat: 1.5 <br /> Price: 5410 <br /> Clarity: I1","Carat: 1.52 <br /> Price: 10308 <br /> Clarity: SI2","Carat: 0.97 <br /> Price: 4063 <br /> Clarity: SI2","Carat: 0.77 <br /> Price: 3321 <br /> Clarity: SI1","Carat: 0.72 <br /> Price: 2708 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 1034 <br /> Clarity: VVS2","Carat: 0.41 <br /> Price: 791 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 4924 <br /> Clarity: SI1","Carat: 1.03 <br /> Price: 9205 <br /> Clarity: VVS2","Carat: 1.74 <br /> Price: 8731 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 6666 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 552 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 1187 <br /> Clarity: IF","Carat: 0.7 <br /> Price: 1758 <br /> Clarity: SI2","Carat: 1.24 <br /> Price: 6867 <br /> Clarity: SI1","Carat: 1 <br /> Price: 8020 <br /> Clarity: VS1","Carat: 1 <br /> Price: 4569 <br /> Clarity: VS2","Carat: 0.6 <br /> Price: 1433 <br /> Clarity: SI2","Carat: 1.31 <br /> Price: 8922 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 730 <br /> Clarity: VVS1","Carat: 0.7 <br /> Price: 2381 <br /> Clarity: VS1","Carat: 0.38 <br /> Price: 871 <br /> Clarity: VS1","Carat: 1.59 <br /> Price: 7123 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 6244 <br /> Clarity: VS2","Carat: 0.38 <br /> Price: 1193 <br /> Clarity: IF","Carat: 0.55 <br /> Price: 1238 <br /> Clarity: SI2","Carat: 2.02 <br /> Price: 9728 <br /> Clarity: SI2","Carat: 0.24 <br /> Price: 504 <br /> Clarity: VVS1","Carat: 0.34 <br /> Price: 596 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 3752 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 921 <br /> Clarity: IF","Carat: 0.31 <br /> Price: 805 <br /> Clarity: VVS2","Carat: 1 <br /> Price: 3965 <br /> Clarity: SI2","Carat: 0.97 <br /> Price: 2357 <br /> Clarity: I1","Carat: 0.4 <br /> Price: 591 <br /> Clarity: VS2","Carat: 0.62 <br /> Price: 2161 <br /> Clarity: VS1","Carat: 1.03 <br /> Price: 5330 <br /> Clarity: SI1","Carat: 1 <br /> Price: 4737 <br /> Clarity: SI1","Carat: 0.28 <br /> Price: 646 <br /> Clarity: VVS1","Carat: 0.48 <br /> Price: 960 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 5288 <br /> Clarity: SI1","Carat: 1 <br /> Price: 6115 <br /> Clarity: VS1","Carat: 0.43 <br /> Price: 670 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 8688 <br /> Clarity: VVS2","Carat: 0.38 <br /> Price: 879 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 709 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 3107 <br /> Clarity: VS1","Carat: 0.91 <br /> Price: 3751 <br /> Clarity: VS2","Carat: 0.66 <br /> Price: 2178 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 4038 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 589 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 965 <br /> Clarity: VVS2","Carat: 1.56 <br /> Price: 13787 <br /> Clarity: VS2","Carat: 0.79 <br /> Price: 2482 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 4118 <br /> Clarity: SI2","Carat: 0.71 <br /> Price: 2320 <br /> Clarity: VS2","Carat: 2.01 <br /> Price: 8104 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1154 <br /> Clarity: SI2","Carat: 1.04 <br /> Price: 4770 <br /> Clarity: SI1","Carat: 1.21 <br /> Price: 5190 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 830 <br /> Clarity: VS1","Carat: 1.16 <br /> Price: 10082 <br /> Clarity: IF","Carat: 0.32 <br /> Price: 645 <br /> Clarity: VS1","Carat: 0.58 <br /> Price: 1984 <br /> Clarity: VS2","Carat: 0.38 <br /> Price: 1026 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 413 <br /> Clarity: I1","Carat: 0.5 <br /> Price: 1706 <br /> Clarity: IF","Carat: 0.32 <br /> Price: 828 <br /> Clarity: VVS2","Carat: 1.15 <br /> Price: 4877 <br /> Clarity: SI2","Carat: 1.05 <br /> Price: 3293 <br /> Clarity: I1","Carat: 0.54 <br /> Price: 2041 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 447 <br /> Clarity: SI2","Carat: 1.29 <br /> Price: 4045 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 1125 <br /> Clarity: VS1","Carat: 0.33 <br /> Price: 723 <br /> Clarity: VS2","Carat: 1.03 <br /> Price: 5050 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2512 <br /> Clarity: VS1","Carat: 1.2 <br /> Price: 6814 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 573 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 557 <br /> Clarity: SI2","Carat: 0.5 <br /> Price: 1095 <br /> Clarity: SI2","Carat: 0.6 <br /> Price: 1777 <br /> Clarity: SI1","Carat: 0.62 <br /> Price: 1378 <br /> Clarity: VS2","Carat: 1.07 <br /> Price: 7222 <br /> Clarity: VS1","Carat: 1.13 <br /> Price: 5062 <br /> Clarity: SI2","Carat: 0.71 <br /> Price: 1626 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 905 <br /> Clarity: IF","Carat: 0.91 <br /> Price: 4392 <br /> Clarity: SI1","Carat: 1.13 <br /> Price: 4873 <br /> Clarity: SI2","Carat: 0.9 <br /> Price: 4580 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 3072 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 691 <br /> Clarity: VVS1","Carat: 1.69 <br /> Price: 11086 <br /> Clarity: VS2","Carat: 0.58 <br /> Price: 1899 <br /> Clarity: VS1","Carat: 1.59 <br /> Price: 9094 <br /> Clarity: SI2","Carat: 0.28 <br /> Price: 487 <br /> Clarity: VS1","Carat: 0.53 <br /> Price: 1857 <br /> Clarity: VS2","Carat: 0.39 <br /> Price: 1213 <br /> Clarity: IF","Carat: 2.16 <br /> Price: 17891 <br /> Clarity: VVS1","Carat: 0.31 <br /> Price: 802 <br /> Clarity: VS2","Carat: 1.55 <br /> Price: 10546 <br /> Clarity: VS2","Carat: 1.6 <br /> Price: 8138 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 971 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4129 <br /> Clarity: SI2","Carat: 1.03 <br /> Price: 4900 <br /> Clarity: SI1","Carat: 0.92 <br /> Price: 4281 <br /> Clarity: SI1","Carat: 0.62 <br /> Price: 2321 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 432 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 737 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2150 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 5141 <br /> Clarity: VS2","Carat: 0.82 <br /> Price: 2893 <br /> Clarity: SI2","Carat: 0.5 <br /> Price: 2475 <br /> Clarity: VVS2","Carat: 0.92 <br /> Price: 4096 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 872 <br /> Clarity: VS2","Carat: 0.91 <br /> Price: 4279 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 931 <br /> Clarity: VVS2","Carat: 0.4 <br /> Price: 684 <br /> Clarity: SI1","Carat: 0.26 <br /> Price: 724 <br /> Clarity: VVS1","Carat: 1.11 <br /> Price: 7758 <br /> Clarity: VS1","Carat: 2.48 <br /> Price: 12883 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 645 <br /> Clarity: VS2","Carat: 1.09 <br /> Price: 7487 <br /> Clarity: VS1","Carat: 0.42 <br /> Price: 992 <br /> Clarity: VS2","Carat: 2.01 <br /> Price: 11825 <br /> Clarity: SI2","Carat: 2.02 <br /> Price: 15974 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 3585 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 8716 <br /> Clarity: VVS2","Carat: 0.32 <br /> Price: 533 <br /> Clarity: IF","Carat: 0.3 <br /> Price: 384 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2839 <br /> Clarity: VS1","Carat: 0.38 <br /> Price: 733 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 732 <br /> Clarity: SI1","Carat: 2.09 <br /> Price: 15773 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 1356 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 945 <br /> Clarity: VVS2","Carat: 0.9 <br /> Price: 3704 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 1092 <br /> Clarity: I1","Carat: 0.28 <br /> Price: 642 <br /> Clarity: VVS1","Carat: 0.9 <br /> Price: 4435 <br /> Clarity: SI1","Carat: 0.54 <br /> Price: 1786 <br /> Clarity: SI1","Carat: 0.28 <br /> Price: 642 <br /> Clarity: VVS2","Carat: 0.31 <br /> Price: 635 <br /> Clarity: IF","Carat: 0.93 <br /> Price: 5729 <br /> Clarity: SI1","Carat: 2.02 <br /> Price: 13109 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 964 <br /> Clarity: SI1","Carat: 1.37 <br /> Price: 12455 <br /> Clarity: VVS1","Carat: 0.37 <br /> Price: 833 <br /> Clarity: VS2","Carat: 2.11 <br /> Price: 16336 <br /> Clarity: SI2","Carat: 0.23 <br /> Price: 530 <br /> Clarity: VVS2","Carat: 0.51 <br /> Price: 1717 <br /> Clarity: VS1","Carat: 1.09 <br /> Price: 6852 <br /> Clarity: SI2","Carat: 1.18 <br /> Price: 5412 <br /> Clarity: SI1","Carat: 1.17 <br /> Price: 7155 <br /> Clarity: VS1","Carat: 0.67 <br /> Price: 3178 <br /> Clarity: VVS1","Carat: 1.6 <br /> Price: 16240 <br /> Clarity: VS2","Carat: 0.76 <br /> Price: 2036 <br /> Clarity: VS1","Carat: 1.04 <br /> Price: 5561 <br /> Clarity: VS2","Carat: 2.01 <br /> Price: 9793 <br /> Clarity: SI1","Carat: 1.06 <br /> Price: 2239 <br /> Clarity: I1","Carat: 1.24 <br /> Price: 13178 <br /> Clarity: IF","Carat: 1.06 <br /> Price: 4060 <br /> Clarity: SI1","Carat: 1.16 <br /> Price: 4657 <br /> Clarity: SI1","Carat: 1.13 <br /> Price: 7016 <br /> Clarity: VS2","Carat: 0.51 <br /> Price: 1682 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1103 <br /> Clarity: SI1","Carat: 1 <br /> Price: 3276 <br /> Clarity: SI2","Carat: 1.08 <br /> Price: 4953 <br /> Clarity: SI1","Carat: 1.55 <br /> Price: 14386 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 1367 <br /> Clarity: VVS1","Carat: 0.91 <br /> Price: 3800 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 954 <br /> Clarity: VVS1","Carat: 1.07 <br /> Price: 7577 <br /> Clarity: VS2","Carat: 0.57 <br /> Price: 2753 <br /> Clarity: IF","Carat: 1.57 <br /> Price: 15144 <br /> Clarity: VVS2","Carat: 0.71 <br /> Price: 1917 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 675 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 421 <br /> Clarity: SI1","Carat: 0.35 <br /> Price: 503 <br /> Clarity: SI1","Carat: 0.45 <br /> Price: 1817 <br /> Clarity: VVS2","Carat: 0.52 <br /> Price: 1737 <br /> Clarity: VS2","Carat: 0.38 <br /> Price: 1064 <br /> Clarity: VS1","Carat: 1.23 <br /> Price: 6206 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 576 <br /> Clarity: SI2","Carat: 0.51 <br /> Price: 1438 <br /> Clarity: SI1","Carat: 0.39 <br /> Price: 587 <br /> Clarity: SI2","Carat: 0.9 <br /> Price: 3989 <br /> Clarity: VS2","Carat: 1.18 <br /> Price: 12175 <br /> Clarity: VVS1","Carat: 1.53 <br /> Price: 13629 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 886 <br /> Clarity: IF","Carat: 0.76 <br /> Price: 3549 <br /> Clarity: VVS2","Carat: 0.51 <br /> Price: 1569 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 737 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 1153 <br /> Clarity: VS1","Carat: 1.05 <br /> Price: 5352 <br /> Clarity: VVS2","Carat: 0.52 <br /> Price: 2359 <br /> Clarity: VVS2","Carat: 1 <br /> Price: 3790 <br /> Clarity: VS2","Carat: 0.28 <br /> Price: 598 <br /> Clarity: VS2","Carat: 0.23 <br /> Price: 364 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 5215 <br /> Clarity: SI1","Carat: 0.77 <br /> Price: 3345 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 6851 <br /> Clarity: SI1","Carat: 1.7 <br /> Price: 13949 <br /> Clarity: VS1","Carat: 1 <br /> Price: 3752 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 706 <br /> Clarity: VS1","Carat: 1.11 <br /> Price: 4578 <br /> Clarity: SI2","Carat: 0.85 <br /> Price: 3084 <br /> Clarity: SI1","Carat: 1 <br /> Price: 6037 <br /> Clarity: VVS2","Carat: 1.08 <br /> Price: 5256 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 8732 <br /> Clarity: VVS2","Carat: 0.51 <br /> Price: 1402 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 10584 <br /> Clarity: VS1","Carat: 1.26 <br /> Price: 7567 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 4506 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 571 <br /> Clarity: SI1","Carat: 0.53 <br /> Price: 1607 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 1187 <br /> Clarity: VVS2","Carat: 0.53 <br /> Price: 1395 <br /> Clarity: SI1","Carat: 0.43 <br /> Price: 795 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 1436 <br /> Clarity: VS2","Carat: 0.8 <br /> Price: 3296 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 3123 <br /> Clarity: VS1","Carat: 0.34 <br /> Price: 650 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 4693 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1845 <br /> Clarity: VS2","Carat: 1.51 <br /> Price: 9741 <br /> Clarity: VS2","Carat: 2.01 <br /> Price: 18607 <br /> Clarity: VS2","Carat: 1.64 <br /> Price: 5624 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 694 <br /> Clarity: VVS2","Carat: 1.04 <br /> Price: 3832 <br /> Clarity: SI2","Carat: 0.78 <br /> Price: 3105 <br /> Clarity: VS1","Carat: 0.76 <br /> Price: 4229 <br /> Clarity: VS1","Carat: 0.52 <br /> Price: 1833 <br /> Clarity: VVS2","Carat: 0.44 <br /> Price: 1238 <br /> Clarity: VS1","Carat: 1.11 <br /> Price: 4973 <br /> Clarity: SI2","Carat: 0.9 <br /> Price: 3407 <br /> Clarity: SI1","Carat: 0.56 <br /> Price: 1819 <br /> Clarity: VS2","Carat: 0.79 <br /> Price: 4570 <br /> Clarity: VVS1","Carat: 0.71 <br /> Price: 3135 <br /> Clarity: VS2","Carat: 0.73 <br /> Price: 2426 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 667 <br /> Clarity: VS2","Carat: 1.03 <br /> Price: 9691 <br /> Clarity: VVS1","Carat: 0.32 <br /> Price: 505 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 1668 <br /> Clarity: VS1","Carat: 1 <br /> Price: 3998 <br /> Clarity: SI2","Carat: 1.33 <br /> Price: 6316 <br /> Clarity: VS2","Carat: 1.54 <br /> Price: 7589 <br /> Clarity: SI1","Carat: 1.7 <br /> Price: 15568 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 3801 <br /> Clarity: SI2","Carat: 0.57 <br /> Price: 1949 <br /> Clarity: VS1","Carat: 1.21 <br /> Price: 5124 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 494 <br /> Clarity: VVS1","Carat: 1.01 <br /> Price: 3399 <br /> Clarity: SI1","Carat: 1.04 <br /> Price: 5105 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1791 <br /> Clarity: VVS2","Carat: 0.32 <br /> Price: 864 <br /> Clarity: IF","Carat: 1 <br /> Price: 4389 <br /> Clarity: SI1","Carat: 1.3 <br /> Price: 6443 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 593 <br /> Clarity: SI2","Carat: 0.5 <br /> Price: 1583 <br /> Clarity: VS1","Carat: 0.43 <br /> Price: 975 <br /> Clarity: SI1","Carat: 1.24 <br /> Price: 10388 <br /> Clarity: VVS2","Carat: 0.32 <br /> Price: 648 <br /> Clarity: VS2","Carat: 0.53 <br /> Price: 1607 <br /> Clarity: VS2","Carat: 1.15 <br /> Price: 5750 <br /> Clarity: VS2","Carat: 1.21 <br /> Price: 4824 <br /> Clarity: SI2","Carat: 1.04 <br /> Price: 6278 <br /> Clarity: VS2","Carat: 0.27 <br /> Price: 518 <br /> Clarity: VS1","Carat: 1.04 <br /> Price: 9882 <br /> Clarity: VVS2","Carat: 0.71 <br /> Price: 2590 <br /> Clarity: VS1","Carat: 0.27 <br /> Price: 583 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 808 <br /> Clarity: VS2","Carat: 1.5 <br /> Price: 6340 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4513 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 3987 <br /> Clarity: I1","Carat: 0.56 <br /> Price: 1580 <br /> Clarity: SI1","Carat: 2.06 <br /> Price: 14982 <br /> Clarity: SI2","Carat: 1.2 <br /> Price: 6026 <br /> Clarity: SI1","Carat: 0.73 <br /> Price: 2326 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 828 <br /> Clarity: VS2","Carat: 0.73 <br /> Price: 2752 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 3205 <br /> Clarity: SI2","Carat: 0.25 <br /> Price: 558 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 3895 <br /> Clarity: SI2","Carat: 1.09 <br /> Price: 10314 <br /> Clarity: VVS2","Carat: 0.41 <br /> Price: 1259 <br /> Clarity: IF","Carat: 0.31 <br /> Price: 625 <br /> Clarity: VS2","Carat: 0.53 <br /> Price: 2158 <br /> Clarity: VVS2","Carat: 0.52 <br /> Price: 1644 <br /> Clarity: VS2","Carat: 0.58 <br /> Price: 2227 <br /> Clarity: VS1","Carat: 0.38 <br /> Price: 855 <br /> Clarity: SI1","Carat: 0.73 <br /> Price: 2564 <br /> Clarity: SI1","Carat: 0.35 <br /> Price: 906 <br /> Clarity: VS2","Carat: 2.04 <br /> Price: 9905 <br /> Clarity: I1","Carat: 0.32 <br /> Price: 648 <br /> Clarity: VS2","Carat: 0.75 <br /> Price: 2588 <br /> Clarity: VS1","Carat: 0.5 <br /> Price: 1094 <br /> Clarity: SI1","Carat: 0.72 <br /> Price: 3274 <br /> Clarity: VS1","Carat: 0.5 <br /> Price: 1190 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 645 <br /> Clarity: VS2","Carat: 0.53 <br /> Price: 2563 <br /> Clarity: VVS2","Carat: 0.72 <br /> Price: 2819 <br /> Clarity: VS1","Carat: 0.79 <br /> Price: 3710 <br /> Clarity: VS2","Carat: 0.35 <br /> Price: 1024 <br /> Clarity: VVS1","Carat: 0.7 <br /> Price: 1806 <br /> Clarity: SI2","Carat: 0.59 <br /> Price: 1561 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 984 <br /> Clarity: VVS1","Carat: 1.45 <br /> Price: 10800 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 878 <br /> Clarity: VVS2","Carat: 0.32 <br /> Price: 828 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 489 <br /> Clarity: VS2","Carat: 0.45 <br /> Price: 1193 <br /> Clarity: VVS1","Carat: 0.32 <br /> Price: 972 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 1074 <br /> Clarity: VVS2","Carat: 0.7 <br /> Price: 2048 <br /> Clarity: SI1","Carat: 0.96 <br /> Price: 2801 <br /> Clarity: I1","Carat: 1.5 <br /> Price: 15992 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 4475 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 819 <br /> Clarity: VVS1","Carat: 1.2 <br /> Price: 6547 <br /> Clarity: VS2","Carat: 0.43 <br /> Price: 1408 <br /> Clarity: VVS2","Carat: 1.04 <br /> Price: 7632 <br /> Clarity: VS1","Carat: 1.13 <br /> Price: 10396 <br /> Clarity: VVS2","Carat: 1.22 <br /> Price: 8485 <br /> Clarity: VS2","Carat: 1.09 <br /> Price: 7583 <br /> Clarity: VVS2","Carat: 1.29 <br /> Price: 5996 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 737 <br /> Clarity: SI1","Carat: 1 <br /> Price: 4620 <br /> Clarity: SI2","Carat: 0.39 <br /> Price: 1095 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 648 <br /> Clarity: VS1","Carat: 0.9 <br /> Price: 4068 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 6806 <br /> Clarity: VS2","Carat: 0.72 <br /> Price: 2578 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 4044 <br /> Clarity: SI2","Carat: 1.02 <br /> Price: 5312 <br /> Clarity: VS2","Carat: 0.79 <br /> Price: 2898 <br /> Clarity: SI1","Carat: 2.16 <br /> Price: 8709 <br /> Clarity: I1","Carat: 0.37 <br /> Price: 957 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 844 <br /> Clarity: VS2","Carat: 0.42 <br /> Price: 1142 <br /> Clarity: VVS2","Carat: 0.41 <br /> Price: 827 <br /> Clarity: VS2","Carat: 0.56 <br /> Price: 2298 <br /> Clarity: IF","Carat: 1.29 <br /> Price: 6588 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 571 <br /> Clarity: VVS1","Carat: 0.4 <br /> Price: 1059 <br /> Clarity: VS2","Carat: 0.35 <br /> Price: 706 <br /> Clarity: VS2","Carat: 1.22 <br /> Price: 5602 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 890 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 2645 <br /> Clarity: IF","Carat: 1.01 <br /> Price: 4194 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 499 <br /> Clarity: SI1","Carat: 0.92 <br /> Price: 4379 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1098 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 1012 <br /> Clarity: VVS1","Carat: 0.34 <br /> Price: 650 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2247 <br /> Clarity: SI2","Carat: 2.08 <br /> Price: 17916 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 3669 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 775 <br /> Clarity: VVS1","Carat: 0.42 <br /> Price: 1142 <br /> Clarity: VVS2","Carat: 0.51 <br /> Price: 1343 <br /> Clarity: VS2","Carat: 0.45 <br /> Price: 978 <br /> Clarity: VVS1","Carat: 2.12 <br /> Price: 17694 <br /> Clarity: SI2","Carat: 0.26 <br /> Price: 453 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 730 <br /> Clarity: VVS2","Carat: 0.33 <br /> Price: 521 <br /> Clarity: VS2","Carat: 0.77 <br /> Price: 2642 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 1080 <br /> Clarity: IF","Carat: 0.9 <br /> Price: 6892 <br /> Clarity: VVS2","Carat: 1.2 <br /> Price: 4516 <br /> Clarity: SI2","Carat: 1.03 <br /> Price: 5018 <br /> Clarity: SI1","Carat: 0.26 <br /> Price: 599 <br /> Clarity: VVS2","Carat: 0.53 <br /> Price: 1665 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 7665 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 666 <br /> Clarity: VS1","Carat: 0.42 <br /> Price: 1096 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 814 <br /> Clarity: VVS1","Carat: 1 <br /> Price: 5775 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 6335 <br /> Clarity: VS2","Carat: 1.51 <br /> Price: 5919 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 666 <br /> Clarity: VS1","Carat: 1.31 <br /> Price: 6676 <br /> Clarity: SI2","Carat: 0.39 <br /> Price: 958 <br /> Clarity: VVS2","Carat: 0.51 <br /> Price: 1146 <br /> Clarity: SI2","Carat: 0.9 <br /> Price: 7747 <br /> Clarity: IF","Carat: 1.01 <br /> Price: 4559 <br /> Clarity: VS2","Carat: 0.39 <br /> Price: 1008 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 471 <br /> Clarity: VS1","Carat: 1.12 <br /> Price: 5046 <br /> Clarity: SI2","Carat: 0.53 <br /> Price: 1332 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 2033 <br /> Clarity: VS1","Carat: 1.5 <br /> Price: 14527 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 1170 <br /> Clarity: IF","Carat: 1.45 <br /> Price: 6455 <br /> Clarity: SI2","Carat: 0.78 <br /> Price: 3298 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 394 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 868 <br /> Clarity: VVS2","Carat: 1 <br /> Price: 3789 <br /> Clarity: SI2","Carat: 2.1 <br /> Price: 12121 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 6250 <br /> Clarity: SI2","Carat: 1.12 <br /> Price: 8721 <br /> Clarity: VVS2","Carat: 1.26 <br /> Price: 6566 <br /> Clarity: VS1","Carat: 1.6 <br /> Price: 12467 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 975 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 593 <br /> Clarity: SI1","Carat: 0.55 <br /> Price: 1186 <br /> Clarity: SI2","Carat: 1.36 <br /> Price: 5103 <br /> Clarity: SI2","Carat: 0.81 <br /> Price: 2655 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4875 <br /> Clarity: SI2","Carat: 2.05 <br /> Price: 15109 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 2264 <br /> Clarity: SI1","Carat: 0.38 <br /> Price: 1211 <br /> Clarity: VVS1","Carat: 1.82 <br /> Price: 12081 <br /> Clarity: SI2","Carat: 0.5 <br /> Price: 1154 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 645 <br /> Clarity: VS1","Carat: 1.44 <br /> Price: 7580 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 1767 <br /> Clarity: VS2","Carat: 1 <br /> Price: 3562 <br /> Clarity: SI2","Carat: 1.07 <br /> Price: 2375 <br /> Clarity: I1","Carat: 0.8 <br /> Price: 2753 <br /> Clarity: VS2","Carat: 0.72 <br /> Price: 3614 <br /> Clarity: IF","Carat: 2.03 <br /> Price: 16945 <br /> Clarity: VS2","Carat: 1.38 <br /> Price: 13250 <br /> Clarity: VVS1","Carat: 0.4 <br /> Price: 982 <br /> Clarity: VVS2","Carat: 0.74 <br /> Price: 2870 <br /> Clarity: SI1","Carat: 1 <br /> Price: 3472 <br /> Clarity: SI2","Carat: 2.01 <br /> Price: 13940 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 4525 <br /> Clarity: VS2","Carat: 1.54 <br /> Price: 8848 <br /> Clarity: VS1","Carat: 0.26 <br /> Price: 597 <br /> Clarity: VVS1","Carat: 0.44 <br /> Price: 990 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 673 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 732 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1960 <br /> Clarity: VVS2","Carat: 0.59 <br /> Price: 1622 <br /> Clarity: VS2","Carat: 2.05 <br /> Price: 15857 <br /> Clarity: SI2","Carat: 1.02 <br /> Price: 5241 <br /> Clarity: VS2","Carat: 0.98 <br /> Price: 4308 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 900 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2338 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1890 <br /> Clarity: VS1","Carat: 0.37 <br /> Price: 684 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2924 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 758 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 687 <br /> Clarity: SI1","Carat: 0.55 <br /> Price: 1556 <br /> Clarity: VS2","Carat: 0.57 <br /> Price: 1963 <br /> Clarity: VS2","Carat: 0.46 <br /> Price: 1314 <br /> Clarity: VVS2","Carat: 0.55 <br /> Price: 2224 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 4007 <br /> Clarity: IF","Carat: 1.14 <br /> Price: 5291 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 791 <br /> Clarity: SI1","Carat: 1 <br /> Price: 7392 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 526 <br /> Clarity: VS1","Carat: 1.1 <br /> Price: 3942 <br /> Clarity: I1","Carat: 0.5 <br /> Price: 1125 <br /> Clarity: SI1","Carat: 2.01 <br /> Price: 14768 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 951 <br /> Clarity: VS1","Carat: 0.34 <br /> Price: 1174 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 764 <br /> Clarity: VVS1","Carat: 0.34 <br /> Price: 1001 <br /> Clarity: VVS1","Carat: 1.51 <br /> Price: 7891 <br /> Clarity: SI1","Carat: 0.45 <br /> Price: 1154 <br /> Clarity: VS1","Carat: 1.08 <br /> Price: 6168 <br /> Clarity: VS2","Carat: 1.13 <br /> Price: 4959 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 487 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 1894 <br /> Clarity: SI2","Carat: 0.34 <br /> Price: 517 <br /> Clarity: VS2","Carat: 1.14 <br /> Price: 11654 <br /> Clarity: VVS1","Carat: 1.07 <br /> Price: 5393 <br /> Clarity: SI2","Carat: 0.43 <br /> Price: 774 <br /> Clarity: SI1","Carat: 1.2 <br /> Price: 9609 <br /> Clarity: VVS1","Carat: 0.5 <br /> Price: 1624 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 739 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 802 <br /> Clarity: VS1","Carat: 0.57 <br /> Price: 1645 <br /> Clarity: VS2","Carat: 0.93 <br /> Price: 4214 <br /> Clarity: SI2","Carat: 2.03 <br /> Price: 18578 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 5919 <br /> Clarity: VS2","Carat: 1.07 <br /> Price: 4686 <br /> Clarity: VS2","Carat: 0.73 <br /> Price: 2330 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 1147 <br /> Clarity: SI1","Carat: 0.53 <br /> Price: 1428 <br /> Clarity: SI1","Carat: 0.37 <br /> Price: 811 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 2271 <br /> Clarity: SI2","Carat: 1.12 <br /> Price: 10211 <br /> Clarity: VVS2","Carat: 1.51 <br /> Price: 10113 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 540 <br /> Clarity: SI2","Carat: 1.87 <br /> Price: 17761 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 1115 <br /> Clarity: VVS2","Carat: 0.7 <br /> Price: 2726 <br /> Clarity: SI1","Carat: 1.09 <br /> Price: 4584 <br /> Clarity: SI2","Carat: 0.38 <br /> Price: 566 <br /> Clarity: VS2","Carat: 1.24 <br /> Price: 8069 <br /> Clarity: VVS2","Carat: 1.2 <br /> Price: 5458 <br /> Clarity: SI1","Carat: 1.14 <br /> Price: 5925 <br /> Clarity: SI1","Carat: 0.62 <br /> Price: 3968 <br /> Clarity: VVS1","Carat: 0.7 <br /> Price: 2373 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 3852 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 4578 <br /> Clarity: SI1","Carat: 1.1 <br /> Price: 4827 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1746 <br /> Clarity: VS1","Carat: 1.53 <br /> Price: 9596 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 802 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 7001 <br /> Clarity: VS2","Carat: 0.55 <br /> Price: 1584 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 7515 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 828 <br /> Clarity: VS1","Carat: 1.22 <br /> Price: 7372 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 462 <br /> Clarity: SI1","Carat: 1.11 <br /> Price: 4771 <br /> Clarity: SI1","Carat: 1.13 <br /> Price: 4936 <br /> Clarity: SI2","Carat: 0.53 <br /> Price: 1648 <br /> Clarity: VS1","Carat: 1.03 <br /> Price: 4292 <br /> Clarity: VS1","Carat: 1.06 <br /> Price: 5143 <br /> Clarity: SI1","Carat: 1.04 <br /> Price: 3603 <br /> Clarity: SI2","Carat: 0.72 <br /> Price: 3478 <br /> Clarity: VS1","Carat: 1.05 <br /> Price: 4797 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 526 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 3039 <br /> Clarity: VS1","Carat: 1.59 <br /> Price: 8829 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 745 <br /> Clarity: VS2","Carat: 0.73 <br /> Price: 3758 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 994 <br /> Clarity: VVS1","Carat: 0.81 <br /> Price: 2797 <br /> Clarity: SI2","Carat: 0.73 <br /> Price: 3311 <br /> Clarity: VS1","Carat: 1.21 <br /> Price: 7703 <br /> Clarity: SI1","Carat: 1 <br /> Price: 4389 <br /> Clarity: SI1","Carat: 1.07 <br /> Price: 3296 <br /> Clarity: SI2","Carat: 0.52 <br /> Price: 1749 <br /> Clarity: VVS2","Carat: 0.74 <br /> Price: 2906 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 3348 <br /> Clarity: SI2","Carat: 1 <br /> Price: 4312 <br /> Clarity: SI1","Carat: 1.03 <br /> Price: 6056 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 999 <br /> Clarity: VS2","Carat: 0.39 <br /> Price: 651 <br /> Clarity: VS2","Carat: 1.27 <br /> Price: 10389 <br /> Clarity: VVS2","Carat: 0.33 <br /> Price: 814 <br /> Clarity: VVS2","Carat: 0.34 <br /> Price: 686 <br /> Clarity: VVS2","Carat: 0.33 <br /> Price: 743 <br /> Clarity: VVS2","Carat: 1.22 <br /> Price: 5960 <br /> Clarity: VS2","Carat: 0.52 <br /> Price: 1778 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 2891 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 891 <br /> Clarity: IF","Carat: 0.8 <br /> Price: 2550 <br /> Clarity: VS2","Carat: 2.06 <br /> Price: 11854 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 1875 <br /> Clarity: VVS2","Carat: 0.31 <br /> Price: 1046 <br /> Clarity: VVS1","Carat: 0.5 <br /> Price: 1935 <br /> Clarity: VVS2","Carat: 0.32 <br /> Price: 828 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 5443 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 4718 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 765 <br /> Clarity: SI1","Carat: 0.77 <br /> Price: 3291 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 972 <br /> Clarity: VS1","Carat: 1.21 <br /> Price: 8286 <br /> Clarity: VVS1","Carat: 1.47 <br /> Price: 6108 <br /> Clarity: SI2","Carat: 0.72 <br /> Price: 2567 <br /> Clarity: SI1","Carat: 2.42 <br /> Price: 16198 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 5786 <br /> Clarity: VS1","Carat: 0.72 <br /> Price: 3236 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 579 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 2753 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2239 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 917 <br /> Clarity: VVS2","Carat: 0.51 <br /> Price: 1882 <br /> Clarity: VVS2","Carat: 0.5 <br /> Price: 1074 <br /> Clarity: SI2","Carat: 1.59 <br /> Price: 9972 <br /> Clarity: VS2","Carat: 1.2 <br /> Price: 5268 <br /> Clarity: SI2","Carat: 0.61 <br /> Price: 1752 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4887 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2473 <br /> Clarity: VS1","Carat: 1.13 <br /> Price: 6391 <br /> Clarity: SI1","Carat: 2.02 <br /> Price: 15334 <br /> Clarity: SI2","Carat: 1.23 <br /> Price: 8541 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 628 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 2790 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 491 <br /> Clarity: VS2","Carat: 1.17 <br /> Price: 5025 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 612 <br /> Clarity: SI2","Carat: 0.92 <br /> Price: 4482 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 702 <br /> Clarity: VS2","Carat: 2.19 <br /> Price: 13978 <br /> Clarity: SI1","Carat: 0.53 <br /> Price: 2177 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 524 <br /> Clarity: VS2","Carat: 0.77 <br /> Price: 2949 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 1171 <br /> Clarity: IF","Carat: 0.32 <br /> Price: 561 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 895 <br /> Clarity: IF","Carat: 0.96 <br /> Price: 4187 <br /> Clarity: VS2","Carat: 0.42 <br /> Price: 803 <br /> Clarity: SI1","Carat: 0.35 <br /> Price: 984 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 335 <br /> Clarity: SI2","Carat: 0.81 <br /> Price: 3097 <br /> Clarity: SI1","Carat: 0.53 <br /> Price: 2137 <br /> Clarity: IF","Carat: 1.2 <br /> Price: 3505 <br /> Clarity: I1","Carat: 2.01 <br /> Price: 17334 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 1007 <br /> Clarity: VS1","Carat: 1.54 <br /> Price: 7287 <br /> Clarity: SI2","Carat: 0.95 <br /> Price: 3750 <br /> Clarity: SI2","Carat: 1.61 <br /> Price: 10197 <br /> Clarity: SI1","Carat: 1.04 <br /> Price: 6564 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 596 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 524 <br /> Clarity: VS2","Carat: 0.51 <br /> Price: 1656 <br /> Clarity: VS2","Carat: 0.8 <br /> Price: 2856 <br /> Clarity: SI2","Carat: 1.17 <br /> Price: 4914 <br /> Clarity: SI1","Carat: 1 <br /> Price: 3304 <br /> Clarity: SI2","Carat: 0.51 <br /> Price: 1656 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 846 <br /> Clarity: VS2","Carat: 0.41 <br /> Price: 1192 <br /> Clarity: VVS2","Carat: 1.1 <br /> Price: 4497 <br /> Clarity: SI1","Carat: 2.39 <br /> Price: 17920 <br /> Clarity: VS1","Carat: 1.7 <br /> Price: 7414 <br /> Clarity: SI2","Carat: 1.28 <br /> Price: 8895 <br /> Clarity: VS1","Carat: 1 <br /> Price: 4620 <br /> Clarity: SI2","Carat: 0.52 <br /> Price: 1600 <br /> Clarity: VS2","Carat: 2.04 <br /> Price: 18795 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4559 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 2828 <br /> Clarity: SI2","Carat: 1.5 <br /> Price: 6750 <br /> Clarity: SI2","Carat: 0.53 <br /> Price: 1939 <br /> Clarity: VS1","Carat: 2.01 <br /> Price: 16677 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 2233 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 802 <br /> Clarity: VS1","Carat: 0.34 <br /> Price: 540 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 1087 <br /> Clarity: VS2","Carat: 0.36 <br /> Price: 518 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 675 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 1151 <br /> Clarity: VS1","Carat: 0.6 <br /> Price: 2316 <br /> Clarity: VVS2","Carat: 0.33 <br /> Price: 521 <br /> Clarity: VS1","Carat: 0.35 <br /> Price: 1063 <br /> Clarity: VS2","Carat: 0.36 <br /> Price: 794 <br /> Clarity: VVS1","Carat: 1 <br /> Price: 3640 <br /> Clarity: SI2","Carat: 1.05 <br /> Price: 5236 <br /> Clarity: VS2","Carat: 0.41 <br /> Price: 1015 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 421 <br /> Clarity: SI2","Carat: 0.56 <br /> Price: 2010 <br /> Clarity: VS1","Carat: 1.18 <br /> Price: 4746 <br /> Clarity: SI1","Carat: 0.49 <br /> Price: 1011 <br /> Clarity: SI2","Carat: 1.05 <br /> Price: 5468 <br /> Clarity: VS2","Carat: 1.11 <br /> Price: 6982 <br /> Clarity: VS2","Carat: 0.52 <br /> Price: 1689 <br /> Clarity: VS1","Carat: 1.14 <br /> Price: 7275 <br /> Clarity: VS2","Carat: 0.36 <br /> Price: 726 <br /> Clarity: VVS2","Carat: 0.52 <br /> Price: 1170 <br /> Clarity: SI2","Carat: 1.35 <br /> Price: 4423 <br /> Clarity: SI2","Carat: 0.5 <br /> Price: 1138 <br /> Clarity: SI1","Carat: 0.91 <br /> Price: 3180 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 1689 <br /> Clarity: VS1","Carat: 0.62 <br /> Price: 1616 <br /> Clarity: VS2","Carat: 1.51 <br /> Price: 8400 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 3340 <br /> Clarity: VVS2","Carat: 0.4 <br /> Price: 1035 <br /> Clarity: VS2","Carat: 0.48 <br /> Price: 994 <br /> Clarity: SI1","Carat: 0.91 <br /> Price: 4332 <br /> Clarity: SI1","Carat: 1.57 <br /> Price: 10964 <br /> Clarity: VS2","Carat: 0.55 <br /> Price: 1799 <br /> Clarity: VS2","Carat: 1.51 <br /> Price: 11666 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 1066 <br /> Clarity: VVS1","Carat: 1.24 <br /> Price: 11376 <br /> Clarity: IF","Carat: 0.3 <br /> Price: 844 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 628 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 545 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 3214 <br /> Clarity: VS1","Carat: 0.34 <br /> Price: 765 <br /> Clarity: SI1","Carat: 0.22 <br /> Price: 404 <br /> Clarity: VS2","Carat: 1.56 <br /> Price: 8107 <br /> Clarity: VS2","Carat: 0.53 <br /> Price: 1607 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 5174 <br /> Clarity: SI1","Carat: 0.56 <br /> Price: 1743 <br /> Clarity: VVS1","Carat: 1.25 <br /> Price: 5975 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 4642 <br /> Clarity: SI1","Carat: 0.79 <br /> Price: 2878 <br /> Clarity: SI2","Carat: 0.43 <br /> Price: 1056 <br /> Clarity: VVS2","Carat: 0.5 <br /> Price: 1090 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4298 <br /> Clarity: SI2","Carat: 1.59 <br /> Price: 7943 <br /> Clarity: SI1","Carat: 1.2 <br /> Price: 5600 <br /> Clarity: VS1","Carat: 0.28 <br /> Price: 707 <br /> Clarity: VVS1","Carat: 1.09 <br /> Price: 5127 <br /> Clarity: SI2","Carat: 0.76 <br /> Price: 1896 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 4536 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 596 <br /> Clarity: SI2","Carat: 0.54 <br /> Price: 1981 <br /> Clarity: IF","Carat: 1.51 <br /> Price: 13080 <br /> Clarity: VS2","Carat: 0.77 <br /> Price: 3089 <br /> Clarity: SI1","Carat: 0.79 <br /> Price: 2507 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1316 <br /> Clarity: SI1","Carat: 0.23 <br /> Price: 400 <br /> Clarity: VS2","Carat: 0.78 <br /> Price: 2496 <br /> Clarity: SI1","Carat: 0.63 <br /> Price: 1105 <br /> Clarity: SI2","Carat: 0.91 <br /> Price: 3340 <br /> Clarity: VS2","Carat: 0.36 <br /> Price: 810 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 917 <br /> Clarity: VVS2","Carat: 0.31 <br /> Price: 408 <br /> Clarity: SI2","Carat: 0.38 <br /> Price: 815 <br /> Clarity: SI1","Carat: 0.38 <br /> Price: 1176 <br /> Clarity: VVS2","Carat: 1.21 <br /> Price: 5882 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 631 <br /> Clarity: SI1","Carat: 2.03 <br /> Price: 15717 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 6499 <br /> Clarity: VS1","Carat: 0.33 <br /> Price: 666 <br /> Clarity: VS1","Carat: 2.18 <br /> Price: 16690 <br /> Clarity: SI2","Carat: 1.54 <br /> Price: 7486 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 932 <br /> Clarity: VVS2","Carat: 0.33 <br /> Price: 434 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 3177 <br /> Clarity: VS1","Carat: 1.12 <br /> Price: 9214 <br /> Clarity: VVS2","Carat: 0.51 <br /> Price: 3920 <br /> Clarity: VVS2","Carat: 1.16 <br /> Price: 5002 <br /> Clarity: SI1","Carat: 0.91 <br /> Price: 3181 <br /> Clarity: SI2","Carat: 1.5 <br /> Price: 12821 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 1013 <br /> Clarity: VVS2","Carat: 2.01 <br /> Price: 15562 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 666 <br /> Clarity: VS1","Carat: 0.55 <br /> Price: 1702 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 731 <br /> Clarity: VS1","Carat: 0.4 <br /> Price: 765 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 3464 <br /> Clarity: VS1","Carat: 1.5 <br /> Price: 12754 <br /> Clarity: SI1","Carat: 0.91 <br /> Price: 3723 <br /> Clarity: SI2","Carat: 0.5 <br /> Price: 1939 <br /> Clarity: VVS2","Carat: 0.7 <br /> Price: 1867 <br /> Clarity: VS2","Carat: 0.52 <br /> Price: 1111 <br /> Clarity: SI1","Carat: 0.39 <br /> Price: 1154 <br /> Clarity: IF","Carat: 0.28 <br /> Price: 825 <br /> Clarity: IF","Carat: 1.58 <br /> Price: 11526 <br /> Clarity: VVS1","Carat: 1.34 <br /> Price: 5245 <br /> Clarity: SI2","Carat: 0.91 <br /> Price: 2854 <br /> Clarity: VS2","Carat: 0.41 <br /> Price: 827 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 854 <br /> Clarity: VVS2","Carat: 0.56 <br /> Price: 1951 <br /> Clarity: VS1","Carat: 1.05 <br /> Price: 5557 <br /> Clarity: SI1","Carat: 1.03 <br /> Price: 4038 <br /> Clarity: SI2","Carat: 0.53 <br /> Price: 2143 <br /> Clarity: VS1","Carat: 0.93 <br /> Price: 4793 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 1214 <br /> Clarity: IF","Carat: 1 <br /> Price: 6518 <br /> Clarity: VS2","Carat: 1.26 <br /> Price: 10302 <br /> Clarity: VVS2","Carat: 0.42 <br /> Price: 984 <br /> Clarity: VS1","Carat: 1.06 <br /> Price: 8093 <br /> Clarity: VS1","Carat: 0.38 <br /> Price: 855 <br /> Clarity: SI1","Carat: 1.41 <br /> Price: 8686 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 1038 <br /> Clarity: VS2","Carat: 1.51 <br /> Price: 14247 <br /> Clarity: VS2","Carat: 1.52 <br /> Price: 12071 <br /> Clarity: SI1","Carat: 1.08 <br /> Price: 5044 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 13653 <br /> Clarity: VS2","Carat: 0.44 <br /> Price: 1003 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 505 <br /> Clarity: VS2","Carat: 1.27 <br /> Price: 7849 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 684 <br /> Clarity: VVS2","Carat: 1.1 <br /> Price: 8932 <br /> Clarity: VVS2","Carat: 1.54 <br /> Price: 10164 <br /> Clarity: VS1","Carat: 0.9 <br /> Price: 3924 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 877 <br /> Clarity: VS1","Carat: 0.35 <br /> Price: 522 <br /> Clarity: SI1","Carat: 1.09 <br /> Price: 6849 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 2950 <br /> Clarity: VS1","Carat: 0.57 <br /> Price: 2052 <br /> Clarity: VS1","Carat: 1.5 <br /> Price: 12000 <br /> Clarity: SI1","Carat: 2.09 <br /> Price: 11576 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 473 <br /> Clarity: VS1","Carat: 2.01 <br /> Price: 15977 <br /> Clarity: SI2","Carat: 0.5 <br /> Price: 1690 <br /> Clarity: VS2","Carat: 0.41 <br /> Price: 1243 <br /> Clarity: VVS2","Carat: 0.82 <br /> Price: 3795 <br /> Clarity: VS2","Carat: 0.42 <br /> Price: 984 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 2553 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 897 <br /> Clarity: VVS1","Carat: 0.59 <br /> Price: 1709 <br /> Clarity: VVS1","Carat: 0.55 <br /> Price: 1848 <br /> Clarity: VS2","Carat: 1.5 <br /> Price: 8662 <br /> Clarity: VS1","Carat: 1.03 <br /> Price: 5523 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4588 <br /> Clarity: VS1","Carat: 0.35 <br /> Price: 906 <br /> Clarity: VS2","Carat: 0.56 <br /> Price: 1737 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2498 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 3062 <br /> Clarity: SI2","Carat: 0.38 <br /> Price: 633 <br /> Clarity: SI2","Carat: 0.72 <br /> Price: 2949 <br /> Clarity: VS2","Carat: 1.13 <br /> Price: 4766 <br /> Clarity: SI1","Carat: 0.61 <br /> Price: 1805 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 561 <br /> Clarity: VS2","Carat: 0.78 <br /> Price: 2694 <br /> Clarity: VS1","Carat: 1.03 <br /> Price: 4414 <br /> Clarity: SI2","Carat: 0.23 <br /> Price: 530 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 5010 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 2508 <br /> Clarity: SI1","Carat: 0.37 <br /> Price: 957 <br /> Clarity: VS2","Carat: 1.21 <br /> Price: 7513 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1745 <br /> Clarity: VS1","Carat: 0.53 <br /> Price: 1813 <br /> Clarity: VS2","Carat: 2.21 <br /> Price: 16558 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 3011 <br /> Clarity: VS2","Carat: 0.45 <br /> Price: 1027 <br /> Clarity: VS1","Carat: 0.66 <br /> Price: 2618 <br /> Clarity: VS1","Carat: 0.33 <br /> Price: 492 <br /> Clarity: SI2","Carat: 0.52 <br /> Price: 1720 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 456 <br /> Clarity: VS2","Carat: 2.04 <br /> Price: 18127 <br /> Clarity: VS1","Carat: 0.36 <br /> Price: 624 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 616 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 5939 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 6838 <br /> Clarity: SI1","Carat: 0.24 <br /> Price: 478 <br /> Clarity: VVS1","Carat: 0.34 <br /> Price: 517 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 2863 <br /> Clarity: SI1","Carat: 1.07 <br /> Price: 5532 <br /> Clarity: SI1","Carat: 1 <br /> Price: 4788 <br /> Clarity: SI1","Carat: 0.27 <br /> Price: 496 <br /> Clarity: VVS2","Carat: 2.14 <br /> Price: 16390 <br /> Clarity: VS2","Carat: 2.24 <br /> Price: 15852 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2537 <br /> Clarity: SI1","Carat: 1.19 <br /> Price: 10436 <br /> Clarity: VVS2","Carat: 1.5 <br /> Price: 10406 <br /> Clarity: VS1","Carat: 0.56 <br /> Price: 1993 <br /> Clarity: VVS1","Carat: 0.79 <br /> Price: 3436 <br /> Clarity: VS1","Carat: 0.77 <br /> Price: 2753 <br /> Clarity: SI2","Carat: 0.34 <br /> Price: 596 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 4412 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 810 <br /> Clarity: IF","Carat: 0.71 <br /> Price: 2162 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 4751 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 714 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 1758 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 673 <br /> Clarity: VS1","Carat: 1.03 <br /> Price: 4764 <br /> Clarity: SI2","Carat: 0.57 <br /> Price: 1982 <br /> Clarity: VS1","Carat: 1.13 <br /> Price: 11051 <br /> Clarity: VVS1","Carat: 0.71 <br /> Price: 3002 <br /> Clarity: VS2","Carat: 0.62 <br /> Price: 1348 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 918 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 605 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 2313 <br /> Clarity: SI1","Carat: 0.92 <br /> Price: 3744 <br /> Clarity: SI2","Carat: 1.24 <br /> Price: 5412 <br /> Clarity: VS2","Carat: 0.53 <br /> Price: 1163 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 4659 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 654 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 3454 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 7357 <br /> Clarity: VS1","Carat: 0.59 <br /> Price: 1552 <br /> Clarity: VS2","Carat: 0.72 <br /> Price: 4228 <br /> Clarity: IF","Carat: 2.03 <br /> Price: 12714 <br /> Clarity: SI2","Carat: 0.57 <br /> Price: 1292 <br /> Clarity: SI1","Carat: 0.35 <br /> Price: 879 <br /> Clarity: VS2","Carat: 1.06 <br /> Price: 3996 <br /> Clarity: SI2","Carat: 1.21 <br /> Price: 5324 <br /> Clarity: SI2","Carat: 0.76 <br /> Price: 3183 <br /> Clarity: VS2","Carat: 0.42 <br /> Price: 1042 <br /> Clarity: VVS2","Carat: 0.4 <br /> Price: 552 <br /> Clarity: SI1","Carat: 1.1 <br /> Price: 3604 <br /> Clarity: SI2","Carat: 0.77 <br /> Price: 1940 <br /> Clarity: VS2","Carat: 1.2 <br /> Price: 5242 <br /> Clarity: VS1","Carat: 1.34 <br /> Price: 7893 <br /> Clarity: SI1","Carat: 0.6 <br /> Price: 1897 <br /> Clarity: VS2","Carat: 0.77 <br /> Price: 2089 <br /> Clarity: SI1","Carat: 1.52 <br /> Price: 12916 <br /> Clarity: VS1","Carat: 1.23 <br /> Price: 10646 <br /> Clarity: VVS2","Carat: 0.72 <br /> Price: 2430 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 878 <br /> Clarity: VVS2","Carat: 0.42 <br /> Price: 945 <br /> Clarity: SI1","Carat: 1.52 <br /> Price: 14217 <br /> Clarity: VS2","Carat: 2.11 <br /> Price: 17871 <br /> Clarity: SI2","Carat: 0.58 <br /> Price: 1865 <br /> Clarity: VS1","Carat: 0.36 <br /> Price: 568 <br /> Clarity: VS2","Carat: 0.51 <br /> Price: 1343 <br /> Clarity: SI1","Carat: 0.93 <br /> Price: 5171 <br /> Clarity: VS1","Carat: 0.64 <br /> Price: 2420 <br /> Clarity: IF","Carat: 0.94 <br /> Price: 3518 <br /> Clarity: SI2","Carat: 1.7 <br /> Price: 15797 <br /> Clarity: VS2","Carat: 0.25 <br /> Price: 576 <br /> Clarity: VVS2","Carat: 0.32 <br /> Price: 477 <br /> Clarity: SI1","Carat: 0.56 <br /> Price: 1924 <br /> Clarity: VS1","Carat: 0.95 <br /> Price: 4738 <br /> Clarity: SI1","Carat: 0.29 <br /> Price: 674 <br /> Clarity: VVS2","Carat: 0.33 <br /> Price: 1240 <br /> Clarity: IF","Carat: 0.41 <br /> Price: 969 <br /> Clarity: SI1","Carat: 1 <br /> Price: 4144 <br /> Clarity: VS1","Carat: 1.02 <br /> Price: 4832 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 515 <br /> Clarity: SI1","Carat: 2.05 <br /> Price: 7026 <br /> Clarity: I1","Carat: 0.7 <br /> Price: 2854 <br /> Clarity: VS1","Carat: 0.55 <br /> Price: 1744 <br /> Clarity: VS2","Carat: 1.58 <br /> Price: 10546 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4171 <br /> Clarity: SI1","Carat: 2.03 <br /> Price: 12862 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1433 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 730 <br /> Clarity: VVS1","Carat: 0.38 <br /> Price: 933 <br /> Clarity: VS2","Carat: 1.08 <br /> Price: 5455 <br /> Clarity: VS2","Carat: 0.38 <br /> Price: 1117 <br /> Clarity: VS1","Carat: 0.59 <br /> Price: 1698 <br /> Clarity: VVS1","Carat: 0.9 <br /> Price: 2761 <br /> Clarity: VS2","Carat: 1.03 <br /> Price: 10607 <br /> Clarity: VS1","Carat: 0.78 <br /> Price: 2777 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 9408 <br /> Clarity: VS2","Carat: 1.54 <br /> Price: 12196 <br /> Clarity: IF","Carat: 2.05 <br /> Price: 13543 <br /> Clarity: VS2","Carat: 0.53 <br /> Price: 2280 <br /> Clarity: VVS2","Carat: 1.24 <br /> Price: 6319 <br /> Clarity: SI1","Carat: 0.45 <br /> Price: 941 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 1443 <br /> Clarity: SI1","Carat: 0.25 <br /> Price: 548 <br /> Clarity: VVS2","Carat: 0.36 <br /> Price: 713 <br /> Clarity: SI1","Carat: 0.58 <br /> Price: 1666 <br /> Clarity: VS2","Carat: 0.51 <br /> Price: 1656 <br /> Clarity: VS1","Carat: 0.9 <br /> Price: 3377 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2043 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 475 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 1008 <br /> Clarity: VVS1","Carat: 0.41 <br /> Price: 738 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 942 <br /> Clarity: VS2","Carat: 0.53 <br /> Price: 1956 <br /> Clarity: VS2","Carat: 0.25 <br /> Price: 575 <br /> Clarity: VVS2","Carat: 0.55 <br /> Price: 1272 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 1115 <br /> Clarity: VVS2","Carat: 0.7 <br /> Price: 2352 <br /> Clarity: SI1","Carat: 0.73 <br /> Price: 3411 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 657 <br /> Clarity: VS1","Carat: 0.5 <br /> Price: 2025 <br /> Clarity: VVS2","Carat: 0.33 <br /> Price: 727 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 707 <br /> Clarity: VVS2","Carat: 1.51 <br /> Price: 6756 <br /> Clarity: SI1","Carat: 0.8 <br /> Price: 2329 <br /> Clarity: SI2","Carat: 1.41 <br /> Price: 8654 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 765 <br /> Clarity: VS2","Carat: 0.41 <br /> Price: 844 <br /> Clarity: VVS1","Carat: 0.7 <br /> Price: 2889 <br /> Clarity: VS1","Carat: 0.39 <br /> Price: 1008 <br /> Clarity: VVS1","Carat: 0.29 <br /> Price: 669 <br /> Clarity: VVS1","Carat: 2.38 <br /> Price: 16643 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 4309 <br /> Clarity: VS1","Carat: 0.36 <br /> Price: 616 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2936 <br /> Clarity: VVS2","Carat: 0.25 <br /> Price: 438 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4129 <br /> Clarity: VS1","Carat: 1.16 <br /> Price: 5164 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 4189 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 2484 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 473 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 3740 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 945 <br /> Clarity: SI1","Carat: 1.6 <br /> Price: 6899 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 414 <br /> Clarity: SI2","Carat: 0.9 <br /> Price: 3473 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 1949 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 1752 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 382 <br /> Clarity: SI1","Carat: 1 <br /> Price: 5088 <br /> Clarity: SI1","Carat: 0.55 <br /> Price: 1081 <br /> Clarity: SI2","Carat: 0.5 <br /> Price: 1240 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 865 <br /> Clarity: VVS1","Carat: 0.57 <br /> Price: 2008 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4497 <br /> Clarity: SI2","Carat: 1.02 <br /> Price: 4281 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1333 <br /> Clarity: VS2","Carat: 0.25 <br /> Price: 438 <br /> Clarity: VS1","Carat: 0.72 <br /> Price: 2949 <br /> Clarity: VS1","Carat: 1.23 <br /> Price: 6459 <br /> Clarity: VS2","Carat: 1.26 <br /> Price: 7910 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 1712 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2874 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 708 <br /> Clarity: VVS1","Carat: 1.28 <br /> Price: 6726 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 2453 <br /> Clarity: SI2","Carat: 0.53 <br /> Price: 856 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 844 <br /> Clarity: VS2","Carat: 1.88 <br /> Price: 8048 <br /> Clarity: SI2","Carat: 0.9 <br /> Price: 3398 <br /> Clarity: SI2","Carat: 1.52 <br /> Price: 7899 <br /> Clarity: VS2","Carat: 1.4 <br /> Price: 8624 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 494 <br /> Clarity: VVS1","Carat: 1.04 <br /> Price: 4164 <br /> Clarity: SI2","Carat: 0.9 <br /> Price: 3324 <br /> Clarity: SI2","Carat: 2.1 <br /> Price: 12381 <br /> Clarity: SI2","Carat: 2 <br /> Price: 17869 <br /> Clarity: SI1","Carat: 0.36 <br /> Price: 537 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 743 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 645 <br /> Clarity: VS1","Carat: 0.54 <br /> Price: 1662 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 720 <br /> Clarity: SI1","Carat: 1.09 <br /> Price: 2708 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 5430 <br /> Clarity: SI1","Carat: 1.03 <br /> Price: 3530 <br /> Clarity: SI2","Carat: 0.78 <br /> Price: 2200 <br /> Clarity: SI2","Carat: 0.42 <br /> Price: 810 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 805 <br /> Clarity: VS1","Carat: 1.06 <br /> Price: 5293 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 427 <br /> Clarity: SI2","Carat: 0.71 <br /> Price: 2764 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 593 <br /> Clarity: SI1","Carat: 0.37 <br /> Price: 1041 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 880 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 2100 <br /> Clarity: VS1","Carat: 0.35 <br /> Price: 583 <br /> Clarity: SI1","Carat: 0.43 <br /> Price: 1329 <br /> Clarity: VS1","Carat: 0.5 <br /> Price: 1654 <br /> Clarity: VS2","Carat: 1 <br /> Price: 7343 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 3307 <br /> Clarity: SI1","Carat: 0.28 <br /> Price: 646 <br /> Clarity: VVS1","Carat: 0.41 <br /> Price: 930 <br /> Clarity: SI1","Carat: 1.18 <br /> Price: 9137 <br /> Clarity: VVS2","Carat: 1.02 <br /> Price: 4037 <br /> Clarity: SI1","Carat: 1.2 <br /> Price: 10454 <br /> Clarity: VVS2","Carat: 0.74 <br /> Price: 2327 <br /> Clarity: VS2","Carat: 1.2 <br /> Price: 5655 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 3296 <br /> Clarity: VVS2","Carat: 0.53 <br /> Price: 1564 <br /> Clarity: SI1","Carat: 0.26 <br /> Price: 456 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 773 <br /> Clarity: VS2","Carat: 1.15 <br /> Price: 9025 <br /> Clarity: VVS1","Carat: 1.07 <br /> Price: 3296 <br /> Clarity: SI2","Carat: 0.52 <br /> Price: 1619 <br /> Clarity: VS2","Carat: 1.04 <br /> Price: 4484 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 6097 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2826 <br /> Clarity: VS2","Carat: 0.56 <br /> Price: 1340 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 3888 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 905 <br /> Clarity: VVS1","Carat: 1.11 <br /> Price: 7179 <br /> Clarity: VS2","Carat: 1 <br /> Price: 8888 <br /> Clarity: VVS2","Carat: 0.51 <br /> Price: 1268 <br /> Clarity: SI2","Carat: 1.7 <br /> Price: 14611 <br /> Clarity: SI1","Carat: 1.43 <br /> Price: 6727 <br /> Clarity: VS1","Carat: 0.55 <br /> Price: 2303 <br /> Clarity: VVS1","Carat: 0.51 <br /> Price: 1268 <br /> Clarity: SI2","Carat: 1.51 <br /> Price: 5979 <br /> Clarity: I1","Carat: 0.74 <br /> Price: 3030 <br /> Clarity: VS1","Carat: 0.9 <br /> Price: 4297 <br /> Clarity: VS1","Carat: 0.37 <br /> Price: 681 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 6108 <br /> Clarity: VS2","Carat: 1.04 <br /> Price: 3914 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 816 <br /> Clarity: VVS1","Carat: 1.01 <br /> Price: 6105 <br /> Clarity: VS2","Carat: 2.07 <br /> Price: 16337 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 872 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 462 <br /> Clarity: SI1","Carat: 0.8 <br /> Price: 3173 <br /> Clarity: SI2","Carat: 0.72 <br /> Price: 2328 <br /> Clarity: SI2","Carat: 0.82 <br /> Price: 3593 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 1046 <br /> Clarity: IF","Carat: 0.39 <br /> Price: 697 <br /> Clarity: SI1","Carat: 2.08 <br /> Price: 16854 <br /> Clarity: VS2","Carat: 0.42 <br /> Price: 756 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 764 <br /> Clarity: VS1","Carat: 0.36 <br /> Price: 782 <br /> Clarity: VS1","Carat: 0.6 <br /> Price: 806 <br /> Clarity: I1","Carat: 0.9 <br /> Price: 3464 <br /> Clarity: SI2","Carat: 0.74 <br /> Price: 3180 <br /> Clarity: VS2","Carat: 0.74 <br /> Price: 2410 <br /> Clarity: SI2","Carat: 1.48 <br /> Price: 7086 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 652 <br /> Clarity: VS1","Carat: 0.37 <br /> Price: 1082 <br /> Clarity: VVS2","Carat: 0.71 <br /> Price: 2347 <br /> Clarity: VS2","Carat: 0.95 <br /> Price: 3669 <br /> Clarity: VVS2","Carat: 0.73 <br /> Price: 3937 <br /> Clarity: VS1","Carat: 0.37 <br /> Price: 681 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 14179 <br /> Clarity: VS2","Carat: 1.13 <br /> Price: 10228 <br /> Clarity: IF","Carat: 0.4 <br /> Price: 790 <br /> Clarity: VS2","Carat: 1 <br /> Price: 7507 <br /> Clarity: VVS1","Carat: 0.85 <br /> Price: 2442 <br /> Clarity: VS2","Carat: 0.51 <br /> Price: 1800 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 6239 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 10580 <br /> Clarity: VVS2","Carat: 1.54 <br /> Price: 9452 <br /> Clarity: SI2","Carat: 0.92 <br /> Price: 3648 <br /> Clarity: SI2","Carat: 0.76 <br /> Price: 2793 <br /> Clarity: VS2","Carat: 1.04 <br /> Price: 7467 <br /> Clarity: VS1","Carat: 0.9 <br /> Price: 4276 <br /> Clarity: VS2","Carat: 0.37 <br /> Price: 681 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 9043 <br /> Clarity: VVS2","Carat: 1 <br /> Price: 4445 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 863 <br /> Clarity: IF","Carat: 0.36 <br /> Price: 631 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 683 <br /> Clarity: SI2","Carat: 0.71 <br /> Price: 3380 <br /> Clarity: VVS2","Carat: 0.5 <br /> Price: 1420 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 803 <br /> Clarity: SI1","Carat: 1.74 <br /> Price: 12720 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 421 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 741 <br /> Clarity: SI1","Carat: 1.2 <br /> Price: 5610 <br /> Clarity: SI2","Carat: 1.21 <br /> Price: 6397 <br /> Clarity: SI1","Carat: 0.57 <br /> Price: 1741 <br /> Clarity: VS1","Carat: 1.12 <br /> Price: 4478 <br /> Clarity: SI2","Carat: 0.58 <br /> Price: 3090 <br /> Clarity: VVS1","Carat: 0.7 <br /> Price: 2167 <br /> Clarity: SI1","Carat: 1.07 <br /> Price: 5872 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2575 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2138 <br /> Clarity: VS2","Carat: 1.7 <br /> Price: 9380 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 4672 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 743 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 1919 <br /> Clarity: VS2","Carat: 0.27 <br /> Price: 552 <br /> Clarity: VVS2","Carat: 0.4 <br /> Price: 1056 <br /> Clarity: VVS2","Carat: 0.42 <br /> Price: 700 <br /> Clarity: SI1","Carat: 0.25 <br /> Price: 436 <br /> Clarity: VS1","Carat: 0.37 <br /> Price: 1041 <br /> Clarity: VS2","Carat: 1.14 <br /> Price: 7199 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 1085 <br /> Clarity: IF","Carat: 0.34 <br /> Price: 596 <br /> Clarity: SI1","Carat: 0.36 <br /> Price: 636 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 988 <br /> Clarity: IF","Carat: 0.91 <br /> Price: 3921 <br /> Clarity: VS2","Carat: 1.17 <br /> Price: 4619 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 696 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 1114 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 844 <br /> Clarity: VS2","Carat: 0.42 <br /> Price: 773 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 764 <br /> Clarity: VVS1","Carat: 1.01 <br /> Price: 7059 <br /> Clarity: VS1","Carat: 0.4 <br /> Price: 896 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 1007 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 593 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 561 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 4971 <br /> Clarity: SI2","Carat: 2.01 <br /> Price: 11188 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 422 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 372 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2423 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 4819 <br /> Clarity: SI1","Carat: 1.04 <br /> Price: 5152 <br /> Clarity: SI1","Carat: 1.1 <br /> Price: 5359 <br /> Clarity: SI2","Carat: 1.24 <br /> Price: 9738 <br /> Clarity: VS2","Carat: 0.61 <br /> Price: 1659 <br /> Clarity: SI1","Carat: 2.14 <br /> Price: 17513 <br /> Clarity: SI2","Carat: 1.02 <br /> Price: 5565 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 766 <br /> Clarity: VVS2","Carat: 1.15 <br /> Price: 4939 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 855 <br /> Clarity: SI2","Carat: 0.26 <br /> Price: 545 <br /> Clarity: VVS1","Carat: 0.34 <br /> Price: 612 <br /> Clarity: SI2","Carat: 0.8 <br /> Price: 2745 <br /> Clarity: SI1","Carat: 1.21 <br /> Price: 9541 <br /> Clarity: VVS2","Carat: 0.33 <br /> Price: 854 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 802 <br /> Clarity: VS1","Carat: 1.51 <br /> Price: 13646 <br /> Clarity: VS2","Carat: 1.34 <br /> Price: 6904 <br /> Clarity: SI2","Carat: 0.53 <br /> Price: 1435 <br /> Clarity: VVS1","Carat: 1.17 <br /> Price: 8639 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 684 <br /> Clarity: VVS2","Carat: 2 <br /> Price: 16650 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 814 <br /> Clarity: VVS2","Carat: 2.1 <br /> Price: 15035 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 944 <br /> Clarity: IF","Carat: 0.5 <br /> Price: 1614 <br /> Clarity: VS2","Carat: 0.53 <br /> Price: 1435 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 625 <br /> Clarity: VVS2","Carat: 0.8 <br /> Price: 2699 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 828 <br /> Clarity: VS2","Carat: 2.01 <br /> Price: 6686 <br /> Clarity: I1","Carat: 1.11 <br /> Price: 7443 <br /> Clarity: VS2","Carat: 0.36 <br /> Price: 663 <br /> Clarity: SI1","Carat: 0.72 <br /> Price: 3759 <br /> Clarity: VS1","Carat: 1.25 <br /> Price: 5706 <br /> Clarity: SI2","Carat: 1.2 <br /> Price: 8983 <br /> Clarity: VS1","Carat: 0.73 <br /> Price: 2258 <br /> Clarity: VS2","Carat: 1.07 <br /> Price: 4830 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 2085 <br /> Clarity: VS1","Carat: 0.58 <br /> Price: 2233 <br /> Clarity: IF","Carat: 0.73 <br /> Price: 3195 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 9523 <br /> Clarity: SI1","Carat: 0.53 <br /> Price: 1766 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 2234 <br /> Clarity: VS2","Carat: 0.43 <br /> Price: 905 <br /> Clarity: VVS1","Carat: 1.5 <br /> Price: 9009 <br /> Clarity: SI2","Carat: 0.52 <br /> Price: 1694 <br /> Clarity: VS1","Carat: 1.25 <br /> Price: 7246 <br /> Clarity: VS2","Carat: 2.1 <br /> Price: 18648 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 435 <br /> Clarity: VS1","Carat: 0.6 <br /> Price: 2442 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 7805 <br /> Clarity: VVS1","Carat: 0.31 <br /> Price: 872 <br /> Clarity: VS2","Carat: 0.45 <br /> Price: 1254 <br /> Clarity: VVS1","Carat: 0.83 <br /> Price: 2800 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1238 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 836 <br /> Clarity: SI1","Carat: 2.5 <br /> Price: 15246 <br /> Clarity: SI2","Carat: 2 <br /> Price: 13320 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 1949 <br /> Clarity: SI1","Carat: 1.13 <br /> Price: 12549 <br /> Clarity: IF","Carat: 0.31 <br /> Price: 625 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 4749 <br /> Clarity: SI1","Carat: 0.83 <br /> Price: 2259 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 1107 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1410 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 7841 <br /> Clarity: SI2","Carat: 1.3 <br /> Price: 12734 <br /> Clarity: VVS2","Carat: 0.71 <br /> Price: 2839 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 658 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 2629 <br /> Clarity: SI1","Carat: 0.46 <br /> Price: 974 <br /> Clarity: VVS1","Carat: 1.03 <br /> Price: 4473 <br /> Clarity: SI2","Carat: 0.37 <br /> Price: 876 <br /> Clarity: VVS2","Carat: 2.51 <br /> Price: 18255 <br /> Clarity: SI2","Carat: 1.07 <br /> Price: 3783 <br /> Clarity: SI2","Carat: 1 <br /> Price: 6989 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 3295 <br /> Clarity: VVS2","Carat: 0.25 <br /> Price: 576 <br /> Clarity: IF","Carat: 0.34 <br /> Price: 596 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 394 <br /> Clarity: SI2","Carat: 0.55 <br /> Price: 2102 <br /> Clarity: VVS2","Carat: 0.4 <br /> Price: 1053 <br /> Clarity: VS1","Carat: 0.51 <br /> Price: 1304 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 710 <br /> Clarity: VS2","Carat: 0.51 <br /> Price: 1343 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 1365 <br /> Clarity: VVS1","Carat: 0.51 <br /> Price: 1443 <br /> Clarity: SI1","Carat: 1.11 <br /> Price: 5550 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 975 <br /> Clarity: VS2","Carat: 0.43 <br /> Price: 1008 <br /> Clarity: VS1","Carat: 0.36 <br /> Price: 459 <br /> Clarity: VS2","Carat: 1.16 <br /> Price: 6301 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 1594 <br /> Clarity: VS1","Carat: 1.03 <br /> Price: 4520 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 781 <br /> Clarity: VS2","Carat: 0.41 <br /> Price: 711 <br /> Clarity: SI1","Carat: 0.53 <br /> Price: 1812 <br /> Clarity: VS1","Carat: 0.27 <br /> Price: 620 <br /> Clarity: VVS1","Carat: 0.38 <br /> Price: 805 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 447 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 1153 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 830 <br /> Clarity: IF","Carat: 0.3 <br /> Price: 710 <br /> Clarity: VS2","Carat: 0.46 <br /> Price: 1450 <br /> Clarity: IF","Carat: 1.56 <br /> Price: 10739 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 705 <br /> Clarity: SI1","Carat: 0.8 <br /> Price: 3502 <br /> Clarity: VS2","Carat: 0.24 <br /> Price: 417 <br /> Clarity: VS2","Carat: 1.86 <br /> Price: 13420 <br /> Clarity: SI2","Carat: 0.57 <br /> Price: 1244 <br /> Clarity: SI2","Carat: 0.28 <br /> Price: 646 <br /> Clarity: VVS2","Carat: 1.04 <br /> Price: 5381 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 421 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 679 <br /> Clarity: VS1","Carat: 1.1 <br /> Price: 6344 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 4309 <br /> Clarity: VS2","Carat: 0.57 <br /> Price: 1641 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 863 <br /> Clarity: VVS1","Carat: 0.66 <br /> Price: 2017 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 808 <br /> Clarity: VVS2","Carat: 0.52 <br /> Price: 1748 <br /> Clarity: VS1","Carat: 1.34 <br /> Price: 7999 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 489 <br /> Clarity: IF","Carat: 1.22 <br /> Price: 6962 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 673 <br /> Clarity: VS2","Carat: 2.51 <br /> Price: 14543 <br /> Clarity: SI2","Carat: 1.52 <br /> Price: 12707 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 8569 <br /> Clarity: VVS2","Carat: 1 <br /> Price: 4861 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 574 <br /> Clarity: VS1","Carat: 0.52 <br /> Price: 1656 <br /> Clarity: SI1","Carat: 0.46 <br /> Price: 1657 <br /> Clarity: VVS1","Carat: 0.52 <br /> Price: 1289 <br /> Clarity: VS2","Carat: 0.96 <br /> Price: 4355 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 1323 <br /> Clarity: VVS2","Carat: 1.62 <br /> Price: 15188 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 576 <br /> Clarity: SI1","Carat: 1.07 <br /> Price: 4446 <br /> Clarity: SI2","Carat: 1.74 <br /> Price: 15189 <br /> Clarity: VS1","Carat: 0.35 <br /> Price: 669 <br /> Clarity: SI2","Carat: 1.74 <br /> Price: 8896 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 5294 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 6618 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 2255 <br /> Clarity: SI1","Carat: 0.75 <br /> Price: 2484 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 449 <br /> Clarity: VS2","Carat: 0.74 <br /> Price: 2880 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 1986 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 3167 <br /> Clarity: I1","Carat: 2.08 <br /> Price: 15654 <br /> Clarity: VVS1","Carat: 0.34 <br /> Price: 686 <br /> Clarity: VVS2","Carat: 0.75 <br /> Price: 3024 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4355 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 441 <br /> Clarity: SI1","Carat: 1.24 <br /> Price: 7162 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 942 <br /> Clarity: VS2","Carat: 0.69 <br /> Price: 3323 <br /> Clarity: VVS1","Carat: 1.24 <br /> Price: 4791 <br /> Clarity: SI1","Carat: 0.53 <br /> Price: 1046 <br /> Clarity: VS2","Carat: 0.24 <br /> Price: 552 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 4432 <br /> Clarity: SI1","Carat: 1.3 <br /> Price: 10038 <br /> Clarity: VS1","Carat: 1.09 <br /> Price: 5364 <br /> Clarity: SI2","Carat: 0.75 <br /> Price: 2551 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 491 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2047 <br /> Clarity: SI2","Carat: 1.21 <br /> Price: 5556 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 670 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 3135 <br /> Clarity: VS2","Carat: 0.38 <br /> Price: 894 <br /> Clarity: VVS1","Carat: 0.5 <br /> Price: 1407 <br /> Clarity: VS2","Carat: 2.02 <br /> Price: 13564 <br /> Clarity: VS2","Carat: 1.19 <br /> Price: 5698 <br /> Clarity: SI1","Carat: 0.25 <br /> Price: 705 <br /> Clarity: VVS2","Carat: 0.33 <br /> Price: 945 <br /> Clarity: VVS1","Carat: 0.41 <br /> Price: 863 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 803 <br /> Clarity: VVS1","Carat: 0.34 <br /> Price: 813 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 789 <br /> Clarity: VVS1","Carat: 0.41 <br /> Price: 830 <br /> Clarity: VS1","Carat: 0.93 <br /> Price: 3738 <br /> Clarity: SI2","Carat: 0.45 <br /> Price: 1055 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 2161 <br /> Clarity: VS1","Carat: 0.38 <br /> Price: 993 <br /> Clarity: VVS1","Carat: 1.57 <br /> Price: 6629 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 645 <br /> Clarity: VS2","Carat: 1.55 <br /> Price: 10851 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 3095 <br /> Clarity: SI1","Carat: 1.2 <br /> Price: 10053 <br /> Clarity: VVS2","Carat: 0.76 <br /> Price: 2988 <br /> Clarity: IF","Carat: 0.6 <br /> Price: 2347 <br /> Clarity: VVS2","Carat: 0.95 <br /> Price: 3378 <br /> Clarity: SI1","Carat: 0.73 <br /> Price: 2190 <br /> Clarity: SI1","Carat: 2.3 <br /> Price: 16062 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 789 <br /> Clarity: IF","Carat: 0.3 <br /> Price: 673 <br /> Clarity: VS1","Carat: 1.54 <br /> Price: 12171 <br /> Clarity: VVS2","Carat: 0.51 <br /> Price: 1562 <br /> Clarity: VS2","Carat: 0.55 <br /> Price: 1792 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 507 <br /> Clarity: SI1","Carat: 0.26 <br /> Price: 362 <br /> Clarity: SI2","Carat: 0.9 <br /> Price: 4788 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1370 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 6517 <br /> Clarity: VS2","Carat: 0.93 <br /> Price: 4062 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1154 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 600 <br /> Clarity: I1","Carat: 2.01 <br /> Price: 16778 <br /> Clarity: VS2","Carat: 1.1 <br /> Price: 3271 <br /> Clarity: SI2","Carat: 0.54 <br /> Price: 1837 <br /> Clarity: VS2","Carat: 0.41 <br /> Price: 827 <br /> Clarity: VS1","Carat: 1.2 <br /> Price: 8334 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 967 <br /> Clarity: VS2","Carat: 1.7 <br /> Price: 10662 <br /> Clarity: VS2","Carat: 0.77 <br /> Price: 3966 <br /> Clarity: VVS1","Carat: 1.11 <br /> Price: 8184 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 4210 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 6103 <br /> Clarity: VS2","Carat: 2.6 <br /> Price: 17209 <br /> Clarity: SI2","Carat: 0.83 <br /> Price: 1968 <br /> Clarity: SI2","Carat: 1.15 <br /> Price: 8501 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 641 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 880 <br /> Clarity: VS1","Carat: 1.65 <br /> Price: 7903 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 13293 <br /> Clarity: VS1","Carat: 1.26 <br /> Price: 5058 <br /> Clarity: SI2","Carat: 0.37 <br /> Price: 746 <br /> Clarity: VS2","Carat: 0.72 <br /> Price: 1864 <br /> Clarity: SI1","Carat: 0.47 <br /> Price: 1021 <br /> Clarity: SI1","Carat: 0.74 <br /> Price: 2761 <br /> Clarity: VVS2","Carat: 0.7 <br /> Price: 2011 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 816 <br /> Clarity: VVS2","Carat: 0.71 <br /> Price: 2452 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 855 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 613 <br /> Clarity: VVS2","Carat: 0.54 <br /> Price: 1846 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 658 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 702 <br /> Clarity: VS2","Carat: 0.36 <br /> Price: 932 <br /> Clarity: VVS2","Carat: 0.54 <br /> Price: 1892 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 702 <br /> Clarity: VS2","Carat: 1.09 <br /> Price: 6453 <br /> Clarity: VS2","Carat: 1.18 <br /> Price: 6013 <br /> Clarity: SI1","Carat: 0.35 <br /> Price: 1024 <br /> Clarity: VVS1","Carat: 1.29 <br /> Price: 6762 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 783 <br /> Clarity: VVS1","Carat: 0.41 <br /> Price: 683 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 526 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 2569 <br /> Clarity: SI1","Carat: 0.37 <br /> Price: 1124 <br /> Clarity: VS1","Carat: 0.24 <br /> Price: 668 <br /> Clarity: VVS2","Carat: 0.41 <br /> Price: 827 <br /> Clarity: VS2","Carat: 2.04 <br /> Price: 16872 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 813 <br /> Clarity: VVS1","Carat: 0.71 <br /> Price: 2599 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 610 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 2594 <br /> Clarity: VVS2","Carat: 0.51 <br /> Price: 1140 <br /> Clarity: VS2","Carat: 0.56 <br /> Price: 1580 <br /> Clarity: SI1","Carat: 0.27 <br /> Price: 470 <br /> Clarity: VS1","Carat: 0.27 <br /> Price: 432 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4137 <br /> Clarity: SI2","Carat: 2.01 <br /> Price: 12338 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 670 <br /> Clarity: VS2","Carat: 0.54 <br /> Price: 1993 <br /> Clarity: VS2","Carat: 0.39 <br /> Price: 787 <br /> Clarity: VS2","Carat: 0.38 <br /> Price: 973 <br /> Clarity: VVS1","Carat: 0.91 <br /> Price: 3485 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 5701 <br /> Clarity: VS2","Carat: 0.92 <br /> Price: 4562 <br /> Clarity: SI1","Carat: 0.62 <br /> Price: 2288 <br /> Clarity: VS2","Carat: 2.44 <br /> Price: 13027 <br /> Clarity: SI2","Carat: 1 <br /> Price: 7812 <br /> Clarity: VS1","Carat: 0.38 <br /> Price: 700 <br /> Clarity: VS1","Carat: 0.57 <br /> Price: 1760 <br /> Clarity: VS2","Carat: 1 <br /> Price: 4427 <br /> Clarity: SI2","Carat: 1 <br /> Price: 4576 <br /> Clarity: SI2","Carat: 0.51 <br /> Price: 1381 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 815 <br /> Clarity: VVS1","Carat: 0.94 <br /> Price: 5980 <br /> Clarity: VS1","Carat: 0.76 <br /> Price: 2518 <br /> Clarity: SI2","Carat: 1 <br /> Price: 4480 <br /> Clarity: SI2","Carat: 0.93 <br /> Price: 3856 <br /> Clarity: SI1","Carat: 0.84 <br /> Price: 3475 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 803 <br /> Clarity: VVS1","Carat: 0.72 <br /> Price: 2404 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 872 <br /> Clarity: VS2","Carat: 0.23 <br /> Price: 530 <br /> Clarity: VVS2","Carat: 1.16 <br /> Price: 4131 <br /> Clarity: SI2","Carat: 0.5 <br /> Price: 1624 <br /> Clarity: VS1","Carat: 0.5 <br /> Price: 2219 <br /> Clarity: IF","Carat: 1.66 <br /> Price: 10134 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 6019 <br /> Clarity: VS2","Carat: 1.58 <br /> Price: 10738 <br /> Clarity: SI1","Carat: 0.57 <br /> Price: 1571 <br /> Clarity: SI1","Carat: 1.56 <br /> Price: 6996 <br /> Clarity: SI2","Carat: 2 <br /> Price: 12196 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2479 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 810 <br /> Clarity: VS1","Carat: 1.02 <br /> Price: 6169 <br /> Clarity: VS2","Carat: 0.73 <br /> Price: 2393 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 702 <br /> Clarity: VS2","Carat: 0.55 <br /> Price: 2656 <br /> Clarity: VVS2","Carat: 1.03 <br /> Price: 5653 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 1015 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 608 <br /> Clarity: SI1","Carat: 1.35 <br /> Price: 5799 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 9710 <br /> Clarity: SI1","Carat: 0.53 <br /> Price: 1956 <br /> Clarity: VS2","Carat: 1.06 <br /> Price: 7334 <br /> Clarity: VS2","Carat: 1.2 <br /> Price: 6019 <br /> Clarity: SI1","Carat: 1.25 <br /> Price: 10636 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 5497 <br /> Clarity: VS2","Carat: 0.51 <br /> Price: 1882 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2415 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 2807 <br /> Clarity: VS1","Carat: 1.03 <br /> Price: 4282 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 816 <br /> Clarity: VS2","Carat: 0.36 <br /> Price: 794 <br /> Clarity: SI1","Carat: 0.36 <br /> Price: 821 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 918 <br /> Clarity: IF","Carat: 0.3 <br /> Price: 895 <br /> Clarity: IF","Carat: 1.29 <br /> Price: 6473 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 408 <br /> Clarity: SI2","Carat: 1 <br /> Price: 4026 <br /> Clarity: SI2","Carat: 1.04 <br /> Price: 4601 <br /> Clarity: SI1","Carat: 2.02 <br /> Price: 17579 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 1024 <br /> Clarity: VS2","Carat: 0.53 <br /> Price: 2269 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 401 <br /> Clarity: SI2","Carat: 0.53 <br /> Price: 1223 <br /> Clarity: SI2","Carat: 0.55 <br /> Price: 1686 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 618 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 651 <br /> Clarity: VS1","Carat: 0.29 <br /> Price: 664 <br /> Clarity: VVS2","Carat: 0.62 <br /> Price: 1327 <br /> Clarity: SI2","Carat: 1.05 <br /> Price: 6132 <br /> Clarity: VS1","Carat: 1.51 <br /> Price: 8260 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 730 <br /> Clarity: VVS2","Carat: 0.62 <br /> Price: 3499 <br /> Clarity: VVS1","Carat: 0.37 <br /> Price: 746 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 1080 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 863 <br /> Clarity: IF","Carat: 0.42 <br /> Price: 1132 <br /> Clarity: VS1","Carat: 0.57 <br /> Price: 2206 <br /> Clarity: VVS2","Carat: 1.51 <br /> Price: 12644 <br /> Clarity: SI1","Carat: 0.98 <br /> Price: 3895 <br /> Clarity: SI2","Carat: 0.56 <br /> Price: 1814 <br /> Clarity: SI1","Carat: 1.27 <br /> Price: 5606 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 608 <br /> Clarity: VS2","Carat: 1 <br /> Price: 3427 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 544 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 612 <br /> Clarity: SI1","Carat: 1.27 <br /> Price: 5953 <br /> Clarity: SI1","Carat: 0.55 <br /> Price: 1853 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 942 <br /> Clarity: VS1","Carat: 1.16 <br /> Price: 7451 <br /> Clarity: VS2","Carat: 0.39 <br /> Price: 716 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 827 <br /> Clarity: VVS2","Carat: 0.42 <br /> Price: 722 <br /> Clarity: SI1","Carat: 2 <br /> Price: 15510 <br /> Clarity: VS2","Carat: 0.55 <br /> Price: 1741 <br /> Clarity: VVS1","Carat: 1.59 <br /> Price: 7978 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 802 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 2310 <br /> Clarity: I1","Carat: 0.43 <br /> Price: 993 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 464 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 3104 <br /> Clarity: SI2","Carat: 0.42 <br /> Price: 1221 <br /> Clarity: VVS2","Carat: 0.85 <br /> Price: 2295 <br /> Clarity: VS2","Carat: 0.23 <br /> Price: 548 <br /> Clarity: VS2","Carat: 0.8 <br /> Price: 2810 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 391 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 5646 <br /> Clarity: SI1","Carat: 0.59 <br /> Price: 1419 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 1001 <br /> Clarity: I1","Carat: 0.27 <br /> Price: 620 <br /> Clarity: VVS2","Carat: 0.64 <br /> Price: 2587 <br /> Clarity: VVS2","Carat: 1.08 <br /> Price: 5487 <br /> Clarity: SI1","Carat: 0.26 <br /> Price: 648 <br /> Clarity: IF","Carat: 0.43 <br /> Price: 1207 <br /> Clarity: VS1","Carat: 0.33 <br /> Price: 761 <br /> Clarity: VVS2","Carat: 1.23 <br /> Price: 9574 <br /> Clarity: VS1","Carat: 1.51 <br /> Price: 14674 <br /> Clarity: VS1","Carat: 0.94 <br /> Price: 3394 <br /> Clarity: SI2","Carat: 1.23 <br /> Price: 5269 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 675 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 571 <br /> Clarity: SI1","Carat: 2.19 <br /> Price: 18067 <br /> Clarity: SI2","Carat: 0.38 <br /> Price: 727 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 3967 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 862 <br /> Clarity: VVS1","Carat: 0.35 <br /> Price: 842 <br /> Clarity: VS1","Carat: 0.33 <br /> Price: 854 <br /> Clarity: VS1","Carat: 1.51 <br /> Price: 9471 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 505 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1752 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 10283 <br /> Clarity: VVS2","Carat: 0.91 <br /> Price: 7006 <br /> Clarity: VVS2","Carat: 1.51 <br /> Price: 12738 <br /> Clarity: SI1","Carat: 1 <br /> Price: 5967 <br /> Clarity: VS2","Carat: 1.13 <br /> Price: 4282 <br /> Clarity: SI2","Carat: 0.5 <br /> Price: 1665 <br /> Clarity: VS1","Carat: 0.8 <br /> Price: 2484 <br /> Clarity: SI2","Carat: 0.36 <br /> Price: 873 <br /> Clarity: VVS2","Carat: 0.36 <br /> Price: 789 <br /> Clarity: VS2","Carat: 1.25 <br /> Price: 4795 <br /> Clarity: VS2","Carat: 1.58 <br /> Price: 7963 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 756 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 658 <br /> Clarity: VS2","Carat: 0.8 <br /> Price: 2616 <br /> Clarity: VS2","Carat: 0.39 <br /> Price: 956 <br /> Clarity: VS1","Carat: 0.42 <br /> Price: 1136 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 851 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 3064 <br /> Clarity: VS2","Carat: 0.54 <br /> Price: 1983 <br /> Clarity: VVS2","Carat: 0.42 <br /> Price: 1103 <br /> Clarity: VVS2","Carat: 0.74 <br /> Price: 2002 <br /> Clarity: SI2","Carat: 1.02 <br /> Price: 4162 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 734 <br /> Clarity: VS1","Carat: 0.36 <br /> Price: 789 <br /> Clarity: VS2","Carat: 2.02 <br /> Price: 14027 <br /> Clarity: SI2","Carat: 0.91 <br /> Price: 2981 <br /> Clarity: SI2","Carat: 1.57 <br /> Price: 10462 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 965 <br /> Clarity: VVS1","Carat: 1.05 <br /> Price: 3528 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 7094 <br /> Clarity: VS1","Carat: 0.51 <br /> Price: 1322 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 4243 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2312 <br /> Clarity: SI1","Carat: 1.18 <br /> Price: 9537 <br /> Clarity: VVS2","Carat: 1.12 <br /> Price: 5058 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 648 <br /> Clarity: VS2","Carat: 2.13 <br /> Price: 17073 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 506 <br /> Clarity: VS1","Carat: 1.02 <br /> Price: 4718 <br /> Clarity: SI1","Carat: 0.38 <br /> Price: 827 <br /> Clarity: SI2","Carat: 0.72 <br /> Price: 2776 <br /> Clarity: VS2","Carat: 2.01 <br /> Price: 13669 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 791 <br /> Clarity: SI1","Carat: 1.25 <br /> Price: 5932 <br /> Clarity: SI2","Carat: 1.04 <br /> Price: 8341 <br /> Clarity: VS1","Carat: 1.18 <br /> Price: 6960 <br /> Clarity: SI1","Carat: 1 <br /> Price: 4032 <br /> Clarity: VS1","Carat: 0.72 <br /> Price: 2444 <br /> Clarity: VS2","Carat: 1.2 <br /> Price: 6793 <br /> Clarity: VS2","Carat: 0.55 <br /> Price: 1871 <br /> Clarity: VS2","Carat: 1 <br /> Price: 5174 <br /> Clarity: SI1","Carat: 1.05 <br /> Price: 4466 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 7207 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 462 <br /> Clarity: SI1","Carat: 0.75 <br /> Price: 5288 <br /> Clarity: VVS1","Carat: 0.56 <br /> Price: 2016 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4129 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 1079 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 2106 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 3923 <br /> Clarity: SI2","Carat: 0.71 <br /> Price: 2960 <br /> Clarity: VVS2","Carat: 1.2 <br /> Price: 6534 <br /> Clarity: SI1","Carat: 1 <br /> Price: 10562 <br /> Clarity: VVS2","Carat: 1.51 <br /> Price: 9474 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 3437 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 9089 <br /> Clarity: VVS2","Carat: 0.43 <br /> Price: 1169 <br /> Clarity: VVS1","Carat: 0.46 <br /> Price: 1326 <br /> Clarity: VS2","Carat: 1.5 <br /> Price: 7774 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 1085 <br /> Clarity: VVS2","Carat: 0.42 <br /> Price: 847 <br /> Clarity: VS2","Carat: 1.51 <br /> Price: 14294 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1190 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 3755 <br /> Clarity: SI2","Carat: 0.75 <br /> Price: 3192 <br /> Clarity: VS2","Carat: 0.53 <br /> Price: 1800 <br /> Clarity: VS1","Carat: 0.8 <br /> Price: 2564 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2042 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 5946 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 3387 <br /> Clarity: SI1","Carat: 0.61 <br /> Price: 1821 <br /> Clarity: VVS2","Carat: 0.55 <br /> Price: 1715 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 698 <br /> Clarity: VS1","Carat: 0.52 <br /> Price: 1586 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 890 <br /> Clarity: VVS1","Carat: 0.74 <br /> Price: 2960 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 612 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 653 <br /> Clarity: VS1","Carat: 0.56 <br /> Price: 1206 <br /> Clarity: SI2","Carat: 0.36 <br /> Price: 852 <br /> Clarity: VS1","Carat: 0.5 <br /> Price: 1624 <br /> Clarity: VS2","Carat: 1.8 <br /> Price: 14968 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 3026 <br /> Clarity: VS2","Carat: 0.52 <br /> Price: 1625 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 5698 <br /> Clarity: SI1","Carat: 0.53 <br /> Price: 1607 <br /> Clarity: VS2","Carat: 2 <br /> Price: 18440 <br /> Clarity: SI1","Carat: 2.16 <br /> Price: 15714 <br /> Clarity: VS1","Carat: 0.53 <br /> Price: 1610 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 900 <br /> Clarity: VS2","Carat: 0.36 <br /> Price: 1056 <br /> Clarity: VVS1","Carat: 0.84 <br /> Price: 3535 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2674 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 755 <br /> Clarity: VVS2","Carat: 0.7 <br /> Price: 1974 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 990 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 4478 <br /> Clarity: SI2","Carat: 1 <br /> Price: 5151 <br /> Clarity: SI1","Carat: 1.33 <br /> Price: 6963 <br /> Clarity: VS1","Carat: 1 <br /> Price: 3405 <br /> Clarity: I1","Carat: 1.08 <br /> Price: 4933 <br /> Clarity: SI1","Carat: 0.53 <br /> Price: 1020 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 526 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1700 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 6271 <br /> Clarity: VS2","Carat: 0.8 <br /> Price: 2481 <br /> Clarity: SI2","Carat: 0.54 <br /> Price: 1637 <br /> Clarity: VS2","Carat: 0.81 <br /> Price: 2977 <br /> Clarity: SI1","Carat: 0.56 <br /> Price: 1664 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 10888 <br /> Clarity: VVS1","Carat: 0.32 <br /> Price: 449 <br /> Clarity: SI2","Carat: 0.54 <br /> Price: 1240 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 880 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 789 <br /> Clarity: VVS1","Carat: 0.4 <br /> Price: 1111 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 4749 <br /> Clarity: SI1","Carat: 1.17 <br /> Price: 7548 <br /> Clarity: VS2","Carat: 0.35 <br /> Price: 827 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 523 <br /> Clarity: VS1","Carat: 1.51 <br /> Price: 7553 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 997 <br /> Clarity: VVS1","Carat: 1.6 <br /> Price: 11796 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 571 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 802 <br /> Clarity: VS2","Carat: 0.99 <br /> Price: 6094 <br /> Clarity: VS2","Carat: 2.01 <br /> Price: 17849 <br /> Clarity: SI1","Carat: 0.27 <br /> Price: 470 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 450 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 1273 <br /> Clarity: I1","Carat: 0.67 <br /> Price: 2309 <br /> Clarity: VS2","Carat: 0.24 <br /> Price: 608 <br /> Clarity: VVS1","Carat: 0.71 <br /> Price: 2711 <br /> Clarity: VS2","Carat: 0.54 <br /> Price: 1065 <br /> Clarity: SI2","Carat: 0.71 <br /> Price: 2331 <br /> Clarity: SI1","Carat: 2.09 <br /> Price: 13500 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 900 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 3484 <br /> Clarity: I1","Carat: 1.06 <br /> Price: 5247 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 2382 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 705 <br /> Clarity: VVS1","Carat: 2.1 <br /> Price: 13182 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 1744 <br /> Clarity: VS1","Carat: 0.54 <br /> Price: 1985 <br /> Clarity: VVS2","Carat: 1.03 <br /> Price: 5526 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 7534 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 939 <br /> Clarity: VVS1","Carat: 0.41 <br /> Price: 830 <br /> Clarity: VS1","Carat: 0.46 <br /> Price: 1401 <br /> Clarity: VS1","Carat: 1.04 <br /> Price: 4368 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 608 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 1781 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 537 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 680 <br /> Clarity: VS2","Carat: 1.23 <br /> Price: 6088 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 852 <br /> Clarity: VVS1","Carat: 0.34 <br /> Price: 928 <br /> Clarity: VVS2","Carat: 0.31 <br /> Price: 864 <br /> Clarity: VVS2","Carat: 0.65 <br /> Price: 1814 <br /> Clarity: SI1","Carat: 1.58 <br /> Price: 12459 <br /> Clarity: VVS2","Carat: 0.23 <br /> Price: 548 <br /> Clarity: VS1","Carat: 2.02 <br /> Price: 13499 <br /> Clarity: VS2","Carat: 1.83 <br /> Price: 13254 <br /> Clarity: SI2","Carat: 0.91 <br /> Price: 3096 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 802 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 4517 <br /> Clarity: SI2","Carat: 1 <br /> Price: 5600 <br /> Clarity: VS2","Carat: 0.36 <br /> Price: 729 <br /> Clarity: VS2","Carat: 1.51 <br /> Price: 8836 <br /> Clarity: SI1","Carat: 1.34 <br /> Price: 6814 <br /> Clarity: SI1","Carat: 0.92 <br /> Price: 3941 <br /> Clarity: SI2","Carat: 1.16 <br /> Price: 5799 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 857 <br /> Clarity: VVS1","Carat: 1.01 <br /> Price: 4844 <br /> Clarity: SI1","Carat: 2.15 <br /> Price: 5430 <br /> Clarity: I1","Carat: 0.9 <br /> Price: 3948 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 3946 <br /> Clarity: VVS2","Carat: 1.33 <br /> Price: 5437 <br /> Clarity: SI2","Carat: 0.54 <br /> Price: 1384 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 868 <br /> Clarity: VVS1","Carat: 0.31 <br /> Price: 625 <br /> Clarity: VS2","Carat: 1.2 <br /> Price: 7603 <br /> Clarity: VS2","Carat: 0.41 <br /> Price: 1399 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 1211 <br /> Clarity: VVS2","Carat: 1.85 <br /> Price: 5688 <br /> Clarity: I1","Carat: 0.32 <br /> Price: 936 <br /> Clarity: VVS1","Carat: 1.32 <br /> Price: 7143 <br /> Clarity: VS1","Carat: 0.76 <br /> Price: 3671 <br /> Clarity: VVS1","Carat: 0.23 <br /> Price: 499 <br /> Clarity: VVS2","Carat: 0.54 <br /> Price: 1250 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 576 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 4762 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1610 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 4688 <br /> Clarity: SI1","Carat: 0.44 <br /> Price: 694 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 3761 <br /> Clarity: SI2","Carat: 1.23 <br /> Price: 5938 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 2680 <br /> Clarity: SI2","Carat: 1.53 <br /> Price: 9555 <br /> Clarity: VVS1","Carat: 1.3 <br /> Price: 6406 <br /> Clarity: SI2","Carat: 0.73 <br /> Price: 2446 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 435 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 956 <br /> Clarity: VS2","Carat: 0.27 <br /> Price: 480 <br /> Clarity: VVS2","Carat: 1.63 <br /> Price: 9556 <br /> Clarity: SI2","Carat: 0.5 <br /> Price: 1569 <br /> Clarity: VS1","Carat: 0.74 <br /> Price: 2805 <br /> Clarity: VS2","Carat: 1.16 <br /> Price: 7003 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 450 <br /> Clarity: SI1","Carat: 2 <br /> Price: 11189 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 776 <br /> Clarity: VS1","Carat: 1.32 <br /> Price: 7173 <br /> Clarity: SI2","Carat: 0.56 <br /> Price: 1963 <br /> Clarity: VS2","Carat: 0.38 <br /> Price: 603 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 864 <br /> Clarity: IF","Carat: 1.12 <br /> Price: 3864 <br /> Clarity: SI2","Carat: 1.2 <br /> Price: 4973 <br /> Clarity: SI2","Carat: 0.53 <br /> Price: 1830 <br /> Clarity: VS1","Carat: 0.37 <br /> Price: 757 <br /> Clarity: VS2","Carat: 1.61 <br /> Price: 14833 <br /> Clarity: VS2","Carat: 0.79 <br /> Price: 2945 <br /> Clarity: VS2","Carat: 1.51 <br /> Price: 13248 <br /> Clarity: VS2","Carat: 1.04 <br /> Price: 4181 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 5973 <br /> Clarity: SI1","Carat: 1.53 <br /> Price: 10947 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 776 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 473 <br /> Clarity: VS1","Carat: 1.03 <br /> Price: 17590 <br /> Clarity: IF","Carat: 0.3 <br /> Price: 675 <br /> Clarity: VS2","Carat: 1.05 <br /> Price: 7010 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 583 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 464 <br /> Clarity: VVS1","Carat: 0.31 <br /> Price: 593 <br /> Clarity: SI2","Carat: 0.36 <br /> Price: 648 <br /> Clarity: VS2","Carat: 0.51 <br /> Price: 1161 <br /> Clarity: SI1","Carat: 0.75 <br /> Price: 3053 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 3290 <br /> Clarity: IF","Carat: 0.38 <br /> Price: 701 <br /> Clarity: SI1","Carat: 0.43 <br /> Price: 968 <br /> Clarity: VS2","Carat: 1.06 <br /> Price: 3544 <br /> Clarity: I1","Carat: 0.72 <br /> Price: 2585 <br /> Clarity: VS2","Carat: 1.25 <br /> Price: 5264 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 3570 <br /> Clarity: SI1","Carat: 0.53 <br /> Price: 1721 <br /> Clarity: VS1","Carat: 1.21 <br /> Price: 2396 <br /> Clarity: I1","Carat: 0.83 <br /> Price: 3506 <br /> Clarity: SI1","Carat: 1.52 <br /> Price: 9576 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 405 <br /> Clarity: SI2","Carat: 1.5 <br /> Price: 9768 <br /> Clarity: SI1","Carat: 1.21 <br /> Price: 13661 <br /> Clarity: VVS2","Carat: 0.77 <br /> Price: 2939 <br /> Clarity: SI1","Carat: 0.39 <br /> Price: 921 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 837 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 828 <br /> Clarity: VS1","Carat: 0.92 <br /> Price: 3345 <br /> Clarity: SI1","Carat: 0.36 <br /> Price: 568 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 4341 <br /> Clarity: SI2","Carat: 0.25 <br /> Price: 582 <br /> Clarity: IF","Carat: 0.3 <br /> Price: 790 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 665 <br /> Clarity: VVS1","Carat: 1.5 <br /> Price: 7518 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 7466 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 3959 <br /> Clarity: SI1","Carat: 1.06 <br /> Price: 6663 <br /> Clarity: VS1","Carat: 0.9 <br /> Price: 4662 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2394 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1410 <br /> Clarity: SI1","Carat: 1.59 <br /> Price: 8975 <br /> Clarity: SI2","Carat: 1.18 <br /> Price: 3925 <br /> Clarity: SI2","Carat: 1.11 <br /> Price: 4315 <br /> Clarity: VS2","Carat: 1.04 <br /> Price: 4200 <br /> Clarity: SI2","Carat: 0.74 <br /> Price: 2328 <br /> Clarity: SI2","Carat: 0.28 <br /> Price: 642 <br /> Clarity: VVS1","Carat: 1.08 <br /> Price: 5171 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 1621 <br /> Clarity: VS2","Carat: 0.36 <br /> Price: 1043 <br /> Clarity: VVS1","Carat: 1.03 <br /> Price: 4586 <br /> Clarity: SI2","Carat: 0.36 <br /> Price: 1148 <br /> Clarity: IF","Carat: 2 <br /> Price: 11106 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2285 <br /> Clarity: SI1","Carat: 0.77 <br /> Price: 3124 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 1564 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 816 <br /> Clarity: VVS1","Carat: 0.35 <br /> Price: 706 <br /> Clarity: VS1","Carat: 0.42 <br /> Price: 958 <br /> Clarity: VS2","Carat: 0.67 <br /> Price: 1910 <br /> Clarity: SI1","Carat: 0.56 <br /> Price: 1698 <br /> Clarity: VS2","Carat: 2 <br /> Price: 13043 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 3024 <br /> Clarity: SI2","Carat: 0.42 <br /> Price: 1235 <br /> Clarity: VS1","Carat: 0.33 <br /> Price: 797 <br /> Clarity: VS1","Carat: 0.6 <br /> Price: 2112 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 1153 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4690 <br /> Clarity: SI2","Carat: 0.83 <br /> Price: 3249 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 608 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 596 <br /> Clarity: SI1","Carat: 0.35 <br /> Price: 788 <br /> Clarity: VS1","Carat: 0.76 <br /> Price: 2578 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 3445 <br /> Clarity: SI1","Carat: 2 <br /> Price: 15218 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 15688 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2058 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 853 <br /> Clarity: VVS1","Carat: 0.7 <br /> Price: 1953 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4899 <br /> Clarity: SI1","Carat: 1.13 <br /> Price: 13111 <br /> Clarity: VVS2","Carat: 1.52 <br /> Price: 8280 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 728 <br /> Clarity: VVS2","Carat: 0.31 <br /> Price: 761 <br /> Clarity: IF","Carat: 1.09 <br /> Price: 5125 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 1244 <br /> Clarity: SI1","Carat: 0.39 <br /> Price: 853 <br /> Clarity: VVS2","Carat: 1.39 <br /> Price: 5021 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 1177 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2234 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 6288 <br /> Clarity: VS2","Carat: 1.1 <br /> Price: 3745 <br /> Clarity: I1","Carat: 0.32 <br /> Price: 972 <br /> Clarity: VS2","Carat: 0.35 <br /> Price: 614 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 837 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 658 <br /> Clarity: VS2","Carat: 1.52 <br /> Price: 14066 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 4466 <br /> Clarity: SI1","Carat: 0.37 <br /> Price: 624 <br /> Clarity: SI2","Carat: 0.71 <br /> Price: 2564 <br /> Clarity: SI2","Carat: 1.64 <br /> Price: 8743 <br /> Clarity: SI2","Carat: 0.59 <br /> Price: 1623 <br /> Clarity: VS2","Carat: 1.09 <br /> Price: 4315 <br /> Clarity: SI2","Carat: 2.01 <br /> Price: 16111 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 717 <br /> Clarity: SI1","Carat: 1.26 <br /> Price: 9158 <br /> Clarity: VVS2","Carat: 0.76 <br /> Price: 2466 <br /> Clarity: SI2","Carat: 0.35 <br /> Price: 851 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 1836 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 13325 <br /> Clarity: VS2","Carat: 0.28 <br /> Price: 487 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 4006 <br /> Clarity: SI1","Carat: 0.96 <br /> Price: 4480 <br /> Clarity: SI1","Carat: 1.03 <br /> Price: 7251 <br /> Clarity: VS1","Carat: 1.51 <br /> Price: 12648 <br /> Clarity: SI1","Carat: 1.1 <br /> Price: 4472 <br /> Clarity: SI1","Carat: 1.09 <br /> Price: 5125 <br /> Clarity: SI1","Carat: 1.03 <br /> Price: 6467 <br /> Clarity: VS2","Carat: 1.06 <br /> Price: 4257 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 698 <br /> Clarity: VS1","Carat: 1.53 <br /> Price: 15466 <br /> Clarity: VVS2","Carat: 0.52 <br /> Price: 1822 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2762 <br /> Clarity: VS2","Carat: 1.5 <br /> Price: 11688 <br /> Clarity: SI1","Carat: 1 <br /> Price: 7676 <br /> Clarity: VS1","Carat: 1.07 <br /> Price: 5002 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 593 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 652 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 720 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 645 <br /> Clarity: VS2","Carat: 0.91 <br /> Price: 4109 <br /> Clarity: VS1","Carat: 0.5 <br /> Price: 1577 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 645 <br /> Clarity: VVS2","Carat: 0.37 <br /> Price: 1082 <br /> Clarity: VVS2","Carat: 1.1 <br /> Price: 4725 <br /> Clarity: SI2","Carat: 0.34 <br /> Price: 459 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 2387 <br /> Clarity: VVS2","Carat: 0.4 <br /> Price: 945 <br /> Clarity: SI1","Carat: 1.14 <br /> Price: 8121 <br /> Clarity: VS1","Carat: 1.05 <br /> Price: 4729 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 972 <br /> Clarity: VS2","Carat: 1 <br /> Price: 3136 <br /> Clarity: SI2","Carat: 0.51 <br /> Price: 1387 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 4688 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 900 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 665 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 652 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 2863 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4221 <br /> Clarity: SI2","Carat: 0.39 <br /> Price: 1362 <br /> Clarity: IF","Carat: 1.52 <br /> Price: 9533 <br /> Clarity: SI2","Carat: 0.44 <br /> Price: 1196 <br /> Clarity: VVS2","Carat: 0.54 <br /> Price: 1381 <br /> Clarity: VS1","Carat: 0.72 <br /> Price: 3816 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 5533 <br /> Clarity: SI1","Carat: 1.04 <br /> Price: 9218 <br /> Clarity: VVS2","Carat: 0.9 <br /> Price: 4612 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 473 <br /> Clarity: VS1","Carat: 1.7 <br /> Price: 7273 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 6066 <br /> Clarity: SI1","Carat: 0.93 <br /> Price: 4879 <br /> Clarity: VS1","Carat: 0.5 <br /> Price: 1190 <br /> Clarity: SI1","Carat: 0.25 <br /> Price: 740 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 684 <br /> Clarity: VVS2","Carat: 0.71 <br /> Price: 2275 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 683 <br /> Clarity: SI1","Carat: 1.31 <br /> Price: 8261 <br /> Clarity: VS2","Carat: 1.16 <br /> Price: 7128 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 2511 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 407 <br /> Clarity: SI1","Carat: 1 <br /> Price: 4596 <br /> Clarity: SI1","Carat: 1.13 <br /> Price: 7290 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 596 <br /> Clarity: VS2","Carat: 0.68 <br /> Price: 2040 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 1238 <br /> Clarity: VVS2","Carat: 0.53 <br /> Price: 1607 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2437 <br /> Clarity: VS2","Carat: 0.41 <br /> Price: 906 <br /> Clarity: VS1","Carat: 1.61 <br /> Price: 11192 <br /> Clarity: SI1","Carat: 1.21 <br /> Price: 4879 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 449 <br /> Clarity: VS2","Carat: 0.63 <br /> Price: 2442 <br /> Clarity: VVS2","Carat: 1.13 <br /> Price: 6743 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 2045 <br /> Clarity: SI1","Carat: 0.23 <br /> Price: 465 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 5174 <br /> Clarity: SI1","Carat: 0.81 <br /> Price: 4926 <br /> Clarity: VVS2","Carat: 1 <br /> Price: 6701 <br /> Clarity: VS2","Carat: 1.22 <br /> Price: 9203 <br /> Clarity: VS2","Carat: 1.53 <br /> Price: 12968 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 720 <br /> Clarity: VS1","Carat: 0.33 <br /> Price: 666 <br /> Clarity: VS2","Carat: 0.39 <br /> Price: 511 <br /> Clarity: I1","Carat: 1.08 <br /> Price: 3849 <br /> Clarity: SI2","Carat: 0.92 <br /> Price: 4183 <br /> Clarity: SI1","Carat: 0.72 <br /> Price: 2505 <br /> Clarity: SI1","Carat: 0.54 <br /> Price: 1083 <br /> Clarity: SI2","Carat: 0.73 <br /> Price: 1902 <br /> Clarity: SI1","Carat: 0.36 <br /> Price: 852 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 2215 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2354 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 1033 <br /> Clarity: VS2","Carat: 0.41 <br /> Price: 1026 <br /> Clarity: VVS2","Carat: 0.72 <br /> Price: 2642 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 680 <br /> Clarity: VS2","Carat: 1 <br /> Price: 5351 <br /> Clarity: SI1","Carat: 1.77 <br /> Price: 9704 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 3660 <br /> Clarity: SI2","Carat: 2.01 <br /> Price: 15105 <br /> Clarity: SI1","Carat: 0.54 <br /> Price: 1644 <br /> Clarity: VS2","Carat: 1.21 <br /> Price: 5218 <br /> Clarity: SI2","Carat: 1.7 <br /> Price: 18176 <br /> Clarity: VS2","Carat: 0.44 <br /> Price: 1003 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 3385 <br /> Clarity: SI2","Carat: 1.22 <br /> Price: 7673 <br /> Clarity: VS1","Carat: 1.55 <br /> Price: 11871 <br /> Clarity: SI1","Carat: 0.81 <br /> Price: 3043 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1752 <br /> Clarity: VS2","Carat: 1.3 <br /> Price: 5140 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2184 <br /> Clarity: SI2","Carat: 1.02 <br /> Price: 11765 <br /> Clarity: VVS2","Carat: 0.5 <br /> Price: 1893 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 579 <br /> Clarity: VS2","Carat: 0.41 <br /> Price: 969 <br /> Clarity: SI1","Carat: 0.76 <br /> Price: 2489 <br /> Clarity: SI2","Carat: 0.61 <br /> Price: 1228 <br /> Clarity: SI2","Carat: 1.2 <br /> Price: 4844 <br /> Clarity: VS2","Carat: 0.58 <br /> Price: 1569 <br /> Clarity: SI1","Carat: 0.27 <br /> Price: 620 <br /> Clarity: VVS1","Carat: 1.2 <br /> Price: 6160 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 6239 <br /> Clarity: VS1","Carat: 0.51 <br /> Price: 1656 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 523 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 1017 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 3553 <br /> Clarity: VS2","Carat: 1.56 <br /> Price: 17455 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 876 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 765 <br /> Clarity: SI2","Carat: 2.36 <br /> Price: 17569 <br /> Clarity: SI2","Carat: 1.55 <br /> Price: 6966 <br /> Clarity: SI2","Carat: 0.24 <br /> Price: 526 <br /> Clarity: VVS2","Carat: 0.51 <br /> Price: 1443 <br /> Clarity: VS2","Carat: 1.1 <br /> Price: 11233 <br /> Clarity: IF","Carat: 1.02 <br /> Price: 3455 <br /> Clarity: SI2","Carat: 1 <br /> Price: 4851 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 827 <br /> Clarity: I1","Carat: 1.5 <br /> Price: 15851 <br /> Clarity: VS1","Carat: 0.73 <br /> Price: 2821 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 457 <br /> Clarity: VS1","Carat: 0.56 <br /> Price: 1715 <br /> Clarity: VS1","Carat: 0.38 <br /> Price: 963 <br /> Clarity: VS2","Carat: 0.54 <br /> Price: 1662 <br /> Clarity: SI1","Carat: 0.38 <br /> Price: 954 <br /> Clarity: VVS2","Carat: 0.77 <br /> Price: 3176 <br /> Clarity: VS1","Carat: 0.51 <br /> Price: 1760 <br /> Clarity: VS1","Carat: 0.73 <br /> Price: 2583 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 3190 <br /> Clarity: IF","Carat: 0.42 <br /> Price: 1048 <br /> Clarity: VS1","Carat: 1.51 <br /> Price: 10935 <br /> Clarity: SI2","Carat: 0.52 <br /> Price: 1303 <br /> Clarity: SI1","Carat: 1.55 <br /> Price: 10937 <br /> Clarity: SI2","Carat: 0.76 <br /> Price: 4596 <br /> Clarity: IF","Carat: 1.01 <br /> Price: 6407 <br /> Clarity: VS1","Carat: 0.73 <br /> Price: 3667 <br /> Clarity: VVS1","Carat: 0.91 <br /> Price: 4476 <br /> Clarity: SI1","Carat: 0.39 <br /> Price: 787 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 579 <br /> Clarity: VS2","Carat: 0.37 <br /> Price: 454 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 4989 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 1024 <br /> Clarity: VVS2","Carat: 1.12 <br /> Price: 5291 <br /> Clarity: SI1","Carat: 1.15 <br /> Price: 4830 <br /> Clarity: SI2","Carat: 0.42 <br /> Price: 1031 <br /> Clarity: VS1","Carat: 0.42 <br /> Price: 1289 <br /> Clarity: IF","Carat: 0.3 <br /> Price: 878 <br /> Clarity: VVS2","Carat: 0.38 <br /> Price: 1000 <br /> Clarity: VS1","Carat: 1.2 <br /> Price: 7372 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 773 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 3278 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 2681 <br /> Clarity: SI1","Carat: 2 <br /> Price: 15451 <br /> Clarity: SI2","Carat: 0.48 <br /> Price: 1279 <br /> Clarity: VS2","Carat: 0.29 <br /> Price: 478 <br /> Clarity: VVS1","Carat: 0.38 <br /> Price: 587 <br /> Clarity: SI2","Carat: 0.9 <br /> Price: 4381 <br /> Clarity: VVS1","Carat: 1.3 <br /> Price: 5569 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 521 <br /> Clarity: VS1","Carat: 2.52 <br /> Price: 17522 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 1207 <br /> Clarity: VVS1","Carat: 0.62 <br /> Price: 2406 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 988 <br /> Clarity: IF","Carat: 0.54 <br /> Price: 1662 <br /> Clarity: SI1","Carat: 1.04 <br /> Price: 5067 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 725 <br /> Clarity: VS2","Carat: 0.42 <br /> Price: 1031 <br /> Clarity: VS2","Carat: 0.51 <br /> Price: 1172 <br /> Clarity: SI1","Carat: 0.92 <br /> Price: 4639 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 2211 <br /> Clarity: VVS2","Carat: 0.41 <br /> Price: 1356 <br /> Clarity: VVS1","Carat: 0.7 <br /> Price: 2562 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2617 <br /> Clarity: SI1","Carat: 1.59 <br /> Price: 16599 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 776 <br /> Clarity: VS2","Carat: 1.51 <br /> Price: 7139 <br /> Clarity: SI2","Carat: 1.04 <br /> Price: 4890 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 777 <br /> Clarity: VVS2","Carat: 1.05 <br /> Price: 5697 <br /> Clarity: VS2","Carat: 1 <br /> Price: 4088 <br /> Clarity: SI2","Carat: 2.01 <br /> Price: 16737 <br /> Clarity: SI1","Carat: 0.55 <br /> Price: 3006 <br /> Clarity: VVS1","Carat: 0.71 <br /> Price: 2647 <br /> Clarity: VS2","Carat: 0.23 <br /> Price: 362 <br /> Clarity: VS2","Carat: 1.5 <br /> Price: 13720 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 2368 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 4981 <br /> Clarity: VVS1","Carat: 2 <br /> Price: 17724 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 3175 <br /> Clarity: VS1","Carat: 0.79 <br /> Price: 2095 <br /> Clarity: SI1","Carat: 2.28 <br /> Price: 12907 <br /> Clarity: SI2","Carat: 0.37 <br /> Price: 811 <br /> Clarity: VS2","Carat: 1.27 <br /> Price: 5455 <br /> Clarity: SI1","Carat: 0.36 <br /> Price: 689 <br /> Clarity: SI1","Carat: 1 <br /> Price: 7966 <br /> Clarity: VS1","Carat: 0.82 <br /> Price: 3071 <br /> Clarity: VS1","Carat: 0.42 <br /> Price: 984 <br /> Clarity: VS1","Carat: 0.57 <br /> Price: 1565 <br /> Clarity: VS2","Carat: 1.26 <br /> Price: 4939 <br /> Clarity: SI2","Carat: 1.39 <br /> Price: 8476 <br /> Clarity: SI1","Carat: 1 <br /> Price: 4328 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 3888 <br /> Clarity: SI1","Carat: 1.15 <br /> Price: 4418 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 743 <br /> Clarity: SI1","Carat: 0.24 <br /> Price: 432 <br /> Clarity: VVS1","Carat: 0.7 <br /> Price: 2369 <br /> Clarity: VS2","Carat: 1.34 <br /> Price: 8930 <br /> Clarity: SI1","Carat: 0.37 <br /> Price: 749 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 516 <br /> Clarity: SI1","Carat: 2.2 <br /> Price: 17634 <br /> Clarity: SI2","Carat: 1.08 <br /> Price: 10204 <br /> Clarity: VVS1","Carat: 1 <br /> Price: 4851 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 1595 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 6073 <br /> Clarity: VS2","Carat: 1 <br /> Price: 3878 <br /> Clarity: SI1","Carat: 0.75 <br /> Price: 2753 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 935 <br /> Clarity: VS2","Carat: 0.24 <br /> Price: 521 <br /> Clarity: VVS1","Carat: 1.5 <br /> Price: 9281 <br /> Clarity: SI2","Carat: 1.51 <br /> Price: 7136 <br /> Clarity: SI1","Carat: 0.8 <br /> Price: 3673 <br /> Clarity: VS1","Carat: 1 <br /> Price: 1997 <br /> Clarity: I1","Carat: 0.54 <br /> Price: 2096 <br /> Clarity: IF","Carat: 1.07 <br /> Price: 5167 <br /> Clarity: VS2","Carat: 1.08 <br /> Price: 10313 <br /> Clarity: VVS2","Carat: 1.5 <br /> Price: 16169 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 719 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 8736 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2889 <br /> Clarity: VS2","Carat: 0.37 <br /> Price: 584 <br /> Clarity: VS2","Carat: 0.8 <br /> Price: 3293 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 4319 <br /> Clarity: SI2","Carat: 0.5 <br /> Price: 1838 <br /> Clarity: VVS2","Carat: 0.78 <br /> Price: 2228 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 761 <br /> Clarity: VVS2","Carat: 1.06 <br /> Price: 6296 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1624 <br /> Clarity: VS1","Carat: 1.15 <br /> Price: 4772 <br /> Clarity: SI1","Carat: 0.25 <br /> Price: 548 <br /> Clarity: VVS2","Carat: 2.26 <br /> Price: 18426 <br /> Clarity: SI1","Carat: 1.05 <br /> Price: 5492 <br /> Clarity: VS2","Carat: 0.57 <br /> Price: 1961 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 5898 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 645 <br /> Clarity: VS1","Carat: 0.53 <br /> Price: 1893 <br /> Clarity: VVS2","Carat: 0.91 <br /> Price: 2832 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 1151 <br /> Clarity: VVS1","Carat: 1.23 <br /> Price: 8128 <br /> Clarity: VS2","Carat: 1.31 <br /> Price: 6573 <br /> Clarity: VS1","Carat: 0.9 <br /> Price: 3160 <br /> Clarity: SI2","Carat: 0.42 <br /> Price: 921 <br /> Clarity: VVS2","Carat: 1.13 <br /> Price: 5017 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 495 <br /> Clarity: VS2","Carat: 1.05 <br /> Price: 4937 <br /> Clarity: SI1","Carat: 0.58 <br /> Price: 1439 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 868 <br /> Clarity: VVS1","Carat: 1 <br /> Price: 4702 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 1289 <br /> Clarity: IF","Carat: 0.91 <br /> Price: 5167 <br /> Clarity: VVS2","Carat: 0.32 <br /> Price: 972 <br /> Clarity: VS2","Carat: 0.86 <br /> Price: 4665 <br /> Clarity: VS2","Carat: 1.33 <br /> Price: 7325 <br /> Clarity: VS2","Carat: 1.52 <br /> Price: 3105 <br /> Clarity: I1","Carat: 0.72 <br /> Price: 2115 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2184 <br /> Clarity: SI1","Carat: 0.53 <br /> Price: 1914 <br /> Clarity: VVS2","Carat: 0.5 <br /> Price: 1121 <br /> Clarity: SI2","Carat: 0.42 <br /> Price: 1167 <br /> Clarity: VVS2","Carat: 1.03 <br /> Price: 10032 <br /> Clarity: IF","Carat: 1.16 <br /> Price: 6036 <br /> Clarity: SI1","Carat: 2.04 <br /> Price: 12554 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 1116 <br /> Clarity: VVS1","Carat: 1.01 <br /> Price: 12581 <br /> Clarity: IF","Carat: 0.5 <br /> Price: 1154 <br /> Clarity: SI2","Carat: 1.04 <br /> Price: 9705 <br /> Clarity: VVS2","Carat: 0.75 <br /> Price: 3013 <br /> Clarity: VS2","Carat: 1.26 <br /> Price: 7400 <br /> Clarity: SI1","Carat: 1.06 <br /> Price: 3740 <br /> Clarity: SI2","Carat: 0.34 <br /> Price: 805 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 454 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 4320 <br /> Clarity: SI1","Carat: 0.91 <br /> Price: 4108 <br /> Clarity: SI1","Carat: 0.36 <br /> Price: 644 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 3407 <br /> Clarity: SI1","Carat: 1.2 <br /> Price: 9904 <br /> Clarity: VS1","Carat: 0.9 <br /> Price: 3872 <br /> Clarity: VVS1","Carat: 1.23 <br /> Price: 11382 <br /> Clarity: VVS2","Carat: 0.75 <br /> Price: 2104 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1351 <br /> Clarity: VS2","Carat: 0.53 <br /> Price: 1412 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 394 <br /> Clarity: SI1","Carat: 0.91 <br /> Price: 4566 <br /> Clarity: VS1","Carat: 0.51 <br /> Price: 1628 <br /> Clarity: VS1","Carat: 2 <br /> Price: 11544 <br /> Clarity: SI2","Carat: 0.75 <br /> Price: 3150 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 3350 <br /> Clarity: SI2","Carat: 1.42 <br /> Price: 7942 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 775 <br /> Clarity: VVS1","Carat: 1.3 <br /> Price: 6042 <br /> Clarity: SI2","Carat: 1.51 <br /> Price: 12141 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 6335 <br /> Clarity: VS2","Carat: 1.27 <br /> Price: 5334 <br /> Clarity: SI2","Carat: 2.01 <br /> Price: 16626 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 654 <br /> Clarity: SI1","Carat: 0.27 <br /> Price: 622 <br /> Clarity: VVS2","Carat: 1.03 <br /> Price: 4095 <br /> Clarity: SI2","Carat: 0.73 <br /> Price: 3226 <br /> Clarity: VS1","Carat: 2.02 <br /> Price: 14027 <br /> Clarity: SI2","Carat: 0.38 <br /> Price: 721 <br /> Clarity: VVS2","Carat: 0.36 <br /> Price: 770 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 10012 <br /> Clarity: SI1","Carat: 1 <br /> Price: 5242 <br /> Clarity: VS2","Carat: 1.51 <br /> Price: 12831 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 779 <br /> Clarity: VVS2","Carat: 1.09 <br /> Price: 5950 <br /> Clarity: SI1","Carat: 1.34 <br /> Price: 7305 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 1059 <br /> Clarity: VS2","Carat: 1.04 <br /> Price: 10539 <br /> Clarity: VVS1","Carat: 0.7 <br /> Price: 2644 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 844 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 577 <br /> Clarity: VS2","Carat: 0.76 <br /> Price: 2148 <br /> Clarity: VS1","Carat: 0.27 <br /> Price: 432 <br /> Clarity: VS1","Carat: 0.33 <br /> Price: 666 <br /> Clarity: VS1","Carat: 0.64 <br /> Price: 1991 <br /> Clarity: SI1","Carat: 0.39 <br /> Price: 942 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2686 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 4633 <br /> Clarity: SI2","Carat: 1 <br /> Price: 4195 <br /> Clarity: SI2","Carat: 0.72 <br /> Price: 2883 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2710 <br /> Clarity: VS1","Carat: 1.23 <br /> Price: 6199 <br /> Clarity: SI2","Carat: 0.74 <br /> Price: 2999 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 485 <br /> Clarity: SI2","Carat: 0.42 <br /> Price: 1389 <br /> Clarity: VVS1","Carat: 0.71 <br /> Price: 3701 <br /> Clarity: VS1","Carat: 0.76 <br /> Price: 2897 <br /> Clarity: SI1","Carat: 0.38 <br /> Price: 1097 <br /> Clarity: VVS2","Carat: 0.9 <br /> Price: 3570 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 4471 <br /> Clarity: SI2","Carat: 1.67 <br /> Price: 8013 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 1882 <br /> Clarity: VS1","Carat: 0.9 <br /> Price: 3796 <br /> Clarity: SI2","Carat: 1.2 <br /> Price: 5016 <br /> Clarity: VS1","Carat: 0.9 <br /> Price: 3595 <br /> Clarity: SI2","Carat: 1.53 <br /> Price: 12499 <br /> Clarity: SI1","Carat: 1.11 <br /> Price: 5408 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 947 <br /> Clarity: SI1","Carat: 1.32 <br /> Price: 11584 <br /> Clarity: VS1","Carat: 0.38 <br /> Price: 695 <br /> Clarity: SI1","Carat: 0.35 <br /> Price: 620 <br /> Clarity: VVS2","Carat: 1.52 <br /> Price: 8127 <br /> Clarity: SI2","Carat: 0.9 <br /> Price: 4004 <br /> Clarity: SI1","Carat: 0.24 <br /> Price: 485 <br /> Clarity: VVS1","Carat: 0.75 <br /> Price: 2923 <br /> Clarity: SI1","Carat: 0.91 <br /> Price: 2913 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 982 <br /> Clarity: VVS2","Carat: 0.33 <br /> Price: 434 <br /> Clarity: SI2","Carat: 1.31 <br /> Price: 7415 <br /> Clarity: VS2","Carat: 0.36 <br /> Price: 932 <br /> Clarity: VS1","Carat: 2.01 <br /> Price: 18741 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 492 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 12430 <br /> Clarity: VS1","Carat: 1.25 <br /> Price: 5252 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 3455 <br /> Clarity: SI2","Carat: 1.22 <br /> Price: 9581 <br /> Clarity: VS1","Carat: 0.34 <br /> Price: 537 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 743 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2196 <br /> Clarity: SI1","Carat: 0.77 <br /> Price: 2159 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1624 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 804 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 844 <br /> Clarity: VS2","Carat: 0.56 <br /> Price: 1389 <br /> Clarity: SI2","Carat: 2.02 <br /> Price: 17825 <br /> Clarity: SI2","Carat: 1.02 <br /> Price: 4633 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 6335 <br /> Clarity: VS2","Carat: 1.51 <br /> Price: 9513 <br /> Clarity: VS1","Carat: 2.04 <br /> Price: 16261 <br /> Clarity: SI2","Carat: 0.71 <br /> Price: 1740 <br /> Clarity: SI2","Carat: 1.57 <br /> Price: 6702 <br /> Clarity: SI2","Carat: 0.35 <br /> Price: 1063 <br /> Clarity: VS2","Carat: 0.53 <br /> Price: 1132 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 801 <br /> Clarity: VS2","Carat: 0.78 <br /> Price: 2489 <br /> Clarity: SI1","Carat: 0.53 <br /> Price: 1160 <br /> Clarity: SI2","Carat: 2.1 <br /> Price: 12121 <br /> Clarity: SI1","Carat: 0.82 <br /> Price: 2692 <br /> Clarity: SI2","Carat: 0.71 <br /> Price: 2207 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 863 <br /> Clarity: IF","Carat: 1.05 <br /> Price: 3352 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 3468 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 693 <br /> Clarity: SI1","Carat: 2.01 <br /> Price: 18395 <br /> Clarity: SI2","Carat: 2.01 <br /> Price: 10401 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 3626 <br /> Clarity: SI2","Carat: 0.42 <br /> Price: 1031 <br /> Clarity: VVS2","Carat: 1.02 <br /> Price: 5100 <br /> Clarity: VS2","Carat: 0.93 <br /> Price: 3685 <br /> Clarity: SI1","Carat: 0.81 <br /> Price: 2973 <br /> Clarity: SI1","Carat: 2.61 <br /> Price: 18756 <br /> Clarity: SI2","Carat: 1.18 <br /> Price: 6112 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2475 <br /> Clarity: SI1","Carat: 0.28 <br /> Price: 434 <br /> Clarity: VS1","Carat: 0.39 <br /> Price: 1167 <br /> Clarity: VVS1","Carat: 0.71 <br /> Price: 2258 <br /> Clarity: SI1","Carat: 1.59 <br /> Price: 8994 <br /> Clarity: SI2","Carat: 1.04 <br /> Price: 4724 <br /> Clarity: SI1","Carat: 0.53 <br /> Price: 1427 <br /> Clarity: VS1","Carat: 0.9 <br /> Price: 4514 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 758 <br /> Clarity: VS2","Carat: 1.18 <br /> Price: 6743 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 421 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 628 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 675 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 975 <br /> Clarity: VS2","Carat: 1.54 <br /> Price: 9081 <br /> Clarity: VS2","Carat: 1.11 <br /> Price: 9734 <br /> Clarity: VVS2","Carat: 0.57 <br /> Price: 1949 <br /> Clarity: VS1","Carat: 1.5 <br /> Price: 9960 <br /> Clarity: VS2","Carat: 1.65 <br /> Price: 14593 <br /> Clarity: VS2","Carat: 0.72 <br /> Price: 3264 <br /> Clarity: VS1","Carat: 0.29 <br /> Price: 664 <br /> Clarity: VVS2","Carat: 0.52 <br /> Price: 1200 <br /> Clarity: SI2","Carat: 0.76 <br /> Price: 2175 <br /> Clarity: VS2","Carat: 1.06 <br /> Price: 7706 <br /> Clarity: VS2","Carat: 1.62 <br /> Price: 8183 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 3360 <br /> Clarity: SI1","Carat: 1.21 <br /> Price: 5893 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 1842 <br /> Clarity: VS2","Carat: 1.12 <br /> Price: 6925 <br /> Clarity: VS2","Carat: 0.96 <br /> Price: 4755 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4150 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 532 <br /> Clarity: VVS2","Carat: 0.32 <br /> Price: 900 <br /> Clarity: VS2","Carat: 1.51 <br /> Price: 12219 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 697 <br /> Clarity: VS1","Carat: 1.16 <br /> Price: 10907 <br /> Clarity: VS2","Carat: 0.66 <br /> Price: 3049 <br /> Clarity: VVS2","Carat: 0.79 <br /> Price: 3806 <br /> Clarity: VS1","Carat: 0.59 <br /> Price: 2200 <br /> Clarity: VS2","Carat: 0.57 <br /> Price: 1963 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 3620 <br /> Clarity: VS2","Carat: 1.26 <br /> Price: 10886 <br /> Clarity: VVS2","Carat: 0.44 <br /> Price: 772 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1801 <br /> Clarity: VS1","Carat: 1.5 <br /> Price: 4620 <br /> Clarity: I1","Carat: 0.51 <br /> Price: 1026 <br /> Clarity: SI2","Carat: 1.2 <br /> Price: 5699 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 2428 <br /> Clarity: VVS2","Carat: 0.9 <br /> Price: 3447 <br /> Clarity: SI1","Carat: 1.06 <br /> Price: 5142 <br /> Clarity: SI1","Carat: 1.24 <br /> Price: 5889 <br /> Clarity: SI2","Carat: 0.53 <br /> Price: 1363 <br /> Clarity: SI1","Carat: 0.72 <br /> Price: 2647 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 5132 <br /> Clarity: SI1","Carat: 0.36 <br /> Price: 759 <br /> Clarity: VS2","Carat: 1.14 <br /> Price: 11549 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 844 <br /> Clarity: IF","Carat: 1.58 <br /> Price: 7333 <br /> Clarity: SI1","Carat: 1.11 <br /> Price: 8698 <br /> Clarity: VS2","Carat: 1.21 <br /> Price: 8505 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 675 <br /> Clarity: VS1","Carat: 0.33 <br /> Price: 463 <br /> Clarity: SI2","Carat: 0.81 <br /> Price: 2386 <br /> Clarity: SI2","Carat: 0.39 <br /> Price: 1009 <br /> Clarity: VS2","Carat: 0.51 <br /> Price: 1438 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4965 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 751 <br /> Clarity: SI1","Carat: 0.39 <br /> Price: 951 <br /> Clarity: VS2","Carat: 0.92 <br /> Price: 4232 <br /> Clarity: SI1","Carat: 1 <br /> Price: 5940 <br /> Clarity: VS1","Carat: 1.57 <br /> Price: 8752 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 828 <br /> Clarity: VS1","Carat: 0.91 <br /> Price: 2863 <br /> Clarity: VS2","Carat: 0.44 <br /> Price: 1187 <br /> Clarity: VVS1","Carat: 0.53 <br /> Price: 1881 <br /> Clarity: VVS2","Carat: 0.58 <br /> Price: 1626 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 1020 <br /> Clarity: IF","Carat: 0.7 <br /> Price: 2808 <br /> Clarity: VS1","Carat: 0.38 <br /> Price: 847 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 764 <br /> Clarity: IF","Carat: 0.7 <br /> Price: 3162 <br /> Clarity: VS1","Carat: 0.77 <br /> Price: 3655 <br /> Clarity: VS1","Carat: 2.01 <br /> Price: 11954 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 3435 <br /> Clarity: VS1","Carat: 1.71 <br /> Price: 8540 <br /> Clarity: SI2","Carat: 0.42 <br /> Price: 771 <br /> Clarity: VVS2","Carat: 2.06 <br /> Price: 12677 <br /> Clarity: SI2","Carat: 1.98 <br /> Price: 12923 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 6039 <br /> Clarity: SI1","Carat: 0.54 <br /> Price: 1637 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1615 <br /> Clarity: VS2","Carat: 1.04 <br /> Price: 4486 <br /> Clarity: SI2","Carat: 0.36 <br /> Price: 793 <br /> Clarity: VS1","Carat: 0.61 <br /> Price: 1591 <br /> Clarity: VS2","Carat: 0.24 <br /> Price: 419 <br /> Clarity: VS1","Carat: 0.91 <br /> Price: 4033 <br /> Clarity: VS2","Carat: 1 <br /> Price: 3080 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 766 <br /> Clarity: IF","Carat: 0.31 <br /> Price: 772 <br /> Clarity: VVS1","Carat: 1.73 <br /> Price: 9510 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 741 <br /> Clarity: VVS2","Carat: 0.31 <br /> Price: 571 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 970 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 7220 <br /> Clarity: IF","Carat: 0.52 <br /> Price: 2426 <br /> Clarity: VVS2","Carat: 1 <br /> Price: 11214 <br /> Clarity: IF","Carat: 1.27 <br /> Price: 6031 <br /> Clarity: VS2","Carat: 1.22 <br /> Price: 4562 <br /> Clarity: VS2","Carat: 0.54 <br /> Price: 1989 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 802 <br /> Clarity: VS2","Carat: 0.98 <br /> Price: 2064 <br /> Clarity: I1","Carat: 0.84 <br /> Price: 4869 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 891 <br /> Clarity: IF","Carat: 0.33 <br /> Price: 752 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 491 <br /> Clarity: VS2","Carat: 2.5 <br /> Price: 12071 <br /> Clarity: SI2","Carat: 1.52 <br /> Price: 13161 <br /> Clarity: VS2","Carat: 0.38 <br /> Price: 998 <br /> Clarity: VS1","Carat: 0.75 <br /> Price: 2744 <br /> Clarity: SI1","Carat: 0.43 <br /> Price: 1048 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 3802 <br /> Clarity: SI2","Carat: 0.77 <br /> Price: 2717 <br /> Clarity: SI1","Carat: 0.35 <br /> Price: 748 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 820 <br /> Clarity: VVS1","Carat: 0.5 <br /> Price: 1187 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 7677 <br /> Clarity: VS1","Carat: 0.72 <br /> Price: 4864 <br /> Clarity: VVS1","Carat: 0.41 <br /> Price: 1059 <br /> Clarity: SI1","Carat: 0.38 <br /> Price: 969 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2100 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 1007 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 3280 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 651 <br /> Clarity: SI1","Carat: 1.08 <br /> Price: 4740 <br /> Clarity: SI1","Carat: 2.02 <br /> Price: 12832 <br /> Clarity: VS2","Carat: 1.56 <br /> Price: 9609 <br /> Clarity: VS2","Carat: 0.73 <br /> Price: 2841 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4821 <br /> Clarity: VS2","Carat: 0.8 <br /> Price: 3913 <br /> Clarity: VS2","Carat: 0.35 <br /> Price: 552 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 2329 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 1115 <br /> Clarity: VVS1","Carat: 0.86 <br /> Price: 3478 <br /> Clarity: SI2","Carat: 0.9 <br /> Price: 4447 <br /> Clarity: SI1","Carat: 1.46 <br /> Price: 7931 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 842 <br /> Clarity: VS2","Carat: 1.22 <br /> Price: 8213 <br /> Clarity: VS2","Carat: 0.53 <br /> Price: 1166 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 2777 <br /> Clarity: VS2","Carat: 1.52 <br /> Price: 16779 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 432 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 653 <br /> Clarity: SI2","Carat: 0.42 <br /> Price: 963 <br /> Clarity: VVS2","Carat: 1.41 <br /> Price: 11644 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 895 <br /> Clarity: IF","Carat: 1.51 <br /> Price: 9569 <br /> Clarity: VS1","Carat: 0.5 <br /> Price: 1624 <br /> Clarity: VS2","Carat: 1.68 <br /> Price: 11147 <br /> Clarity: SI2","Carat: 1.53 <br /> Price: 7730 <br /> Clarity: SI1","Carat: 0.54 <br /> Price: 2356 <br /> Clarity: VVS1","Carat: 1.14 <br /> Price: 7079 <br /> Clarity: VS2","Carat: 2.4 <br /> Price: 15824 <br /> Clarity: SI2","Carat: 0.42 <br /> Price: 1307 <br /> Clarity: VVS1","Carat: 1.03 <br /> Price: 6751 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 3871 <br /> Clarity: SI2","Carat: 1.05 <br /> Price: 4742 <br /> Clarity: SI2","Carat: 1 <br /> Price: 4311 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 788 <br /> Clarity: SI1","Carat: 1 <br /> Price: 3136 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2239 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 775 <br /> Clarity: VS1","Carat: 0.54 <br /> Price: 1208 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 6425 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 2699 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 1657 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4504 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 1050 <br /> Clarity: VS1","Carat: 0.58 <br /> Price: 2117 <br /> Clarity: VS1","Carat: 0.38 <br /> Price: 1140 <br /> Clarity: VS1","Carat: 1.08 <br /> Price: 8674 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 702 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 477 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 658 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 875 <br /> Clarity: VVS1","Carat: 1.51 <br /> Price: 9513 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 668 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 2428 <br /> Clarity: VS2","Carat: 0.48 <br /> Price: 841 <br /> Clarity: SI1","Carat: 1.09 <br /> Price: 5042 <br /> Clarity: SI2","Carat: 0.72 <br /> Price: 2359 <br /> Clarity: SI2","Carat: 1.67 <br /> Price: 11146 <br /> Clarity: SI1","Carat: 0.75 <br /> Price: 2467 <br /> Clarity: VS2","Carat: 0.27 <br /> Price: 893 <br /> Clarity: IF","Carat: 0.41 <br /> Price: 1279 <br /> Clarity: VVS1","Carat: 0.55 <br /> Price: 1401 <br /> Clarity: SI2","Carat: 0.46 <br /> Price: 1139 <br /> Clarity: VS1","Carat: 0.34 <br /> Price: 612 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2394 <br /> Clarity: SI2","Carat: 0.54 <br /> Price: 1401 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 5006 <br /> Clarity: SI1","Carat: 2 <br /> Price: 17889 <br /> Clarity: VS1","Carat: 0.4 <br /> Price: 737 <br /> Clarity: VS1","Carat: 1.07 <br /> Price: 5220 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 1343 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 1264 <br /> Clarity: SI2","Carat: 1.51 <br /> Price: 8930 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 6108 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 6968 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 734 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 850 <br /> Clarity: IF","Carat: 0.3 <br /> Price: 526 <br /> Clarity: SI1","Carat: 1 <br /> Price: 5863 <br /> Clarity: VS1","Carat: 0.36 <br /> Price: 631 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 1687 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 4044 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 1966 <br /> Clarity: VS2","Carat: 0.58 <br /> Price: 1442 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 557 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 5593 <br /> Clarity: VS1","Carat: 1 <br /> Price: 4704 <br /> Clarity: VS2","Carat: 0.76 <br /> Price: 2425 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 603 <br /> Clarity: VS1","Carat: 2.01 <br /> Price: 17688 <br /> Clarity: SI2","Carat: 0.9 <br /> Price: 3418 <br /> Clarity: VS1","Carat: 0.35 <br /> Price: 830 <br /> Clarity: VVS2","Carat: 0.52 <br /> Price: 1111 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 507 <br /> Clarity: SI2","Carat: 1.03 <br /> Price: 4582 <br /> Clarity: VS1","Carat: 1.03 <br /> Price: 6662 <br /> Clarity: VS2","Carat: 0.29 <br /> Price: 518 <br /> Clarity: SI1","Carat: 0.36 <br /> Price: 983 <br /> Clarity: VVS2","Carat: 0.42 <br /> Price: 945 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 956 <br /> Clarity: VVS1","Carat: 0.55 <br /> Price: 1271 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1436 <br /> Clarity: VS2","Carat: 1 <br /> Price: 4395 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 942 <br /> Clarity: VS2","Carat: 0.72 <br /> Price: 2482 <br /> Clarity: SI1","Carat: 0.54 <br /> Price: 2271 <br /> Clarity: IF","Carat: 0.3 <br /> Price: 789 <br /> Clarity: VVS2","Carat: 1.56 <br /> Price: 10424 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 4315 <br /> Clarity: SI1","Carat: 1.11 <br /> Price: 4883 <br /> Clarity: SI2","Carat: 0.83 <br /> Price: 2913 <br /> Clarity: SI1","Carat: 0.28 <br /> Price: 492 <br /> Clarity: VVS2","Carat: 0.43 <br /> Price: 679 <br /> Clarity: SI2","Carat: 0.55 <br /> Price: 1975 <br /> Clarity: VS1","Carat: 0.33 <br /> Price: 492 <br /> Clarity: SI2","Carat: 1.02 <br /> Price: 6648 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 7466 <br /> Clarity: VVS2","Carat: 0.57 <br /> Price: 4032 <br /> Clarity: IF","Carat: 1.01 <br /> Price: 5817 <br /> Clarity: VS2","Carat: 0.55 <br /> Price: 2064 <br /> Clarity: VVS2","Carat: 0.33 <br /> Price: 579 <br /> Clarity: VS1","Carat: 0.9 <br /> Price: 2873 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 614 <br /> Clarity: VVS2","Carat: 0.54 <br /> Price: 1637 <br /> Clarity: VS2","Carat: 0.41 <br /> Price: 1115 <br /> Clarity: VVS2","Carat: 1.15 <br /> Price: 7032 <br /> Clarity: VS1","Carat: 1.03 <br /> Price: 6479 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 1107 <br /> Clarity: VS2","Carat: 0.59 <br /> Price: 1757 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 544 <br /> Clarity: VS2","Carat: 1.62 <br /> Price: 14386 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 506 <br /> Clarity: SI2","Carat: 1.37 <br /> Price: 6751 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 4047 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 942 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 561 <br /> Clarity: VS2","Carat: 0.58 <br /> Price: 1809 <br /> Clarity: VS2","Carat: 1.54 <br /> Price: 17029 <br /> Clarity: VS2","Carat: 1.13 <br /> Price: 4584 <br /> Clarity: SI2","Carat: 0.8 <br /> Price: 4229 <br /> Clarity: VVS2","Carat: 0.4 <br /> Price: 918 <br /> Clarity: SI1","Carat: 0.96 <br /> Price: 3215 <br /> Clarity: SI2","Carat: 0.56 <br /> Price: 2157 <br /> Clarity: IF","Carat: 1 <br /> Price: 5505 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1370 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 780 <br /> Clarity: IF","Carat: 0.51 <br /> Price: 1875 <br /> Clarity: VVS2","Carat: 1.5 <br /> Price: 7476 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 5229 <br /> Clarity: SI1","Carat: 0.58 <br /> Price: 1914 <br /> Clarity: VS2","Carat: 0.53 <br /> Price: 984 <br /> Clarity: SI2","Carat: 0.38 <br /> Price: 1026 <br /> Clarity: VS2","Carat: 0.56 <br /> Price: 1436 <br /> Clarity: SI1","Carat: 0.28 <br /> Price: 487 <br /> Clarity: VS1","Carat: 0.53 <br /> Price: 1727 <br /> Clarity: VS1","Carat: 0.86 <br /> Price: 4191 <br /> Clarity: VS2","Carat: 0.59 <br /> Price: 2319 <br /> Clarity: VS1","Carat: 0.27 <br /> Price: 799 <br /> Clarity: VVS2","Carat: 0.73 <br /> Price: 2496 <br /> Clarity: VVS2","Carat: 0.6 <br /> Price: 1738 <br /> Clarity: SI1","Carat: 0.35 <br /> Price: 767 <br /> Clarity: VS2","Carat: 0.57 <br /> Price: 1728 <br /> Clarity: VS2","Carat: 1.5 <br /> Price: 9408 <br /> Clarity: SI2","Carat: 1.13 <br /> Price: 6682 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 9058 <br /> Clarity: SI2","Carat: 0.81 <br /> Price: 4412 <br /> Clarity: VS1","Carat: 0.23 <br /> Price: 485 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 518 <br /> Clarity: VS1","Carat: 0.78 <br /> Price: 3763 <br /> Clarity: VS2","Carat: 0.25 <br /> Price: 490 <br /> Clarity: VVS1","Carat: 1.5 <br /> Price: 10090 <br /> Clarity: SI2","Carat: 0.93 <br /> Price: 3172 <br /> Clarity: SI2","Carat: 0.52 <br /> Price: 2217 <br /> Clarity: VVS2","Carat: 1.23 <br /> Price: 8233 <br /> Clarity: VS2","Carat: 0.73 <br /> Price: 3606 <br /> Clarity: VVS1","Carat: 0.32 <br /> Price: 949 <br /> Clarity: IF","Carat: 0.72 <br /> Price: 2827 <br /> Clarity: VS2","Carat: 1.5 <br /> Price: 6798 <br /> Clarity: SI2","Carat: 1.12 <br /> Price: 3669 <br /> Clarity: SI2","Carat: 1.5 <br /> Price: 8931 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 1103 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 526 <br /> Clarity: VS1","Carat: 0.34 <br /> Price: 469 <br /> Clarity: VS1","Carat: 0.92 <br /> Price: 3749 <br /> Clarity: SI1","Carat: 0.43 <br /> Price: 1827 <br /> Clarity: VVS1","Carat: 1.1 <br /> Price: 5914 <br /> Clarity: VS2","Carat: 0.87 <br /> Price: 2923 <br /> Clarity: SI1","Carat: 0.96 <br /> Price: 3808 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 2387 <br /> Clarity: SI2","Carat: 1.23 <br /> Price: 5717 <br /> Clarity: SI1","Carat: 0.24 <br /> Price: 336 <br /> Clarity: VVS1","Carat: 0.6 <br /> Price: 1875 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 2400 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 658 <br /> Clarity: VS2","Carat: 1.2 <br /> Price: 5941 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 574 <br /> Clarity: SI2","Carat: 1.6 <br /> Price: 7550 <br /> Clarity: SI1","Carat: 1.52 <br /> Price: 11235 <br /> Clarity: VS2","Carat: 1.09 <br /> Price: 5010 <br /> Clarity: SI1","Carat: 1.2 <br /> Price: 4611 <br /> Clarity: VS2","Carat: 1.59 <br /> Price: 8227 <br /> Clarity: VVS2","Carat: 0.24 <br /> Price: 391 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 1079 <br /> Clarity: VS1","Carat: 0.54 <br /> Price: 1454 <br /> Clarity: VS1","Carat: 1.31 <br /> Price: 8774 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 530 <br /> Clarity: SI2","Carat: 0.35 <br /> Price: 945 <br /> Clarity: VVS1","Carat: 0.31 <br /> Price: 944 <br /> Clarity: IF","Carat: 1 <br /> Price: 6296 <br /> Clarity: VVS1","Carat: 1.95 <br /> Price: 14603 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 990 <br /> Clarity: VVS1","Carat: 0.4 <br /> Price: 1172 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 2112 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2822 <br /> Clarity: SI1","Carat: 1.15 <br /> Price: 4186 <br /> Clarity: SI2","Carat: 1.21 <br /> Price: 4754 <br /> Clarity: VS2","Carat: 0.52 <br /> Price: 1088 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2699 <br /> Clarity: SI1","Carat: 0.82 <br /> Price: 3810 <br /> Clarity: SI1","Carat: 0.26 <br /> Price: 597 <br /> Clarity: VVS2","Carat: 0.92 <br /> Price: 4253 <br /> Clarity: SI1","Carat: 1.11 <br /> Price: 6857 <br /> Clarity: VS2","Carat: 0.43 <br /> Price: 708 <br /> Clarity: SI1","Carat: 2.2 <br /> Price: 12747 <br /> Clarity: SI2","Carat: 1.38 <br /> Price: 5912 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 1128 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 7167 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 4807 <br /> Clarity: SI2","Carat: 2.03 <br /> Price: 12677 <br /> Clarity: SI2","Carat: 0.52 <br /> Price: 1878 <br /> Clarity: VVS2","Carat: 1.21 <br /> Price: 4816 <br /> Clarity: VS2","Carat: 0.44 <br /> Price: 1073 <br /> Clarity: VS2","Carat: 1 <br /> Price: 2912 <br /> Clarity: SI2","Carat: 1.04 <br /> Price: 4435 <br /> Clarity: SI2","Carat: 1.29 <br /> Price: 6588 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 755 <br /> Clarity: SI1","Carat: 1.08 <br /> Price: 4318 <br /> Clarity: SI2","Carat: 0.71 <br /> Price: 3381 <br /> Clarity: VS2","Carat: 1.65 <br /> Price: 17425 <br /> Clarity: VS1","Carat: 0.52 <br /> Price: 1689 <br /> Clarity: VS1","Carat: 1.11 <br /> Price: 4854 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 936 <br /> Clarity: VVS2","Carat: 0.74 <br /> Price: 2609 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 731 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 447 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 1004 <br /> Clarity: VS1","Carat: 0.63 <br /> Price: 1921 <br /> Clarity: SI1","Carat: 1 <br /> Price: 6776 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2212 <br /> Clarity: VS2","Carat: 1.16 <br /> Price: 5805 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 952 <br /> Clarity: VS2","Carat: 0.69 <br /> Price: 2227 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4440 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 1114 <br /> Clarity: VVS1","Carat: 0.5 <br /> Price: 1286 <br /> Clarity: SI1","Carat: 1.79 <br /> Price: 18429 <br /> Clarity: VS2","Carat: 1.05 <br /> Price: 8982 <br /> Clarity: VVS2","Carat: 1.09 <br /> Price: 6288 <br /> Clarity: VS2","Carat: 0.79 <br /> Price: 4180 <br /> Clarity: VVS2","Carat: 0.51 <br /> Price: 1717 <br /> Clarity: VS1","Carat: 0.35 <br /> Price: 706 <br /> Clarity: VVS2","Carat: 1.02 <br /> Price: 4233 <br /> Clarity: SI1","Carat: 1.06 <br /> Price: 5319 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 5002 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1569 <br /> Clarity: VVS2","Carat: 1.51 <br /> Price: 14542 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 789 <br /> Clarity: IF","Carat: 0.31 <br /> Price: 625 <br /> Clarity: VS2","Carat: 1.2 <br /> Price: 8032 <br /> Clarity: VS2","Carat: 1.03 <br /> Price: 5241 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 755 <br /> Clarity: SI1","Carat: 1.11 <br /> Price: 5433 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 761 <br /> Clarity: VVS2","Carat: 0.54 <br /> Price: 1392 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 561 <br /> Clarity: VS2","Carat: 1.26 <br /> Price: 9720 <br /> Clarity: VS2","Carat: 0.46 <br /> Price: 1166 <br /> Clarity: VS1","Carat: 0.44 <br /> Price: 777 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 579 <br /> Clarity: SI1","Carat: 0.58 <br /> Price: 1373 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 776 <br /> Clarity: VS1","Carat: 0.5 <br /> Price: 1316 <br /> Clarity: SI1","Carat: 1.15 <br /> Price: 5296 <br /> Clarity: SI1","Carat: 0.76 <br /> Price: 2120 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 11088 <br /> Clarity: VS1","Carat: 0.51 <br /> Price: 1807 <br /> Clarity: VS2","Carat: 2 <br /> Price: 13098 <br /> Clarity: SI2","Carat: 0.72 <br /> Price: 2150 <br /> Clarity: VVS2","Carat: 1.12 <br /> Price: 3266 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2789 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 3176 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 918 <br /> Clarity: IF","Carat: 1.04 <br /> Price: 2801 <br /> Clarity: I1","Carat: 0.81 <br /> Price: 2795 <br /> Clarity: SI2","Carat: 0.44 <br /> Price: 1194 <br /> Clarity: IF","Carat: 0.38 <br /> Price: 1092 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 756 <br /> Clarity: SI1","Carat: 2.03 <br /> Price: 16309 <br /> Clarity: VS2","Carat: 0.39 <br /> Price: 902 <br /> Clarity: VVS2","Carat: 1.34 <br /> Price: 7453 <br /> Clarity: SI2","Carat: 0.77 <br /> Price: 2642 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 3197 <br /> Clarity: VVS1","Carat: 0.33 <br /> Price: 743 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 3453 <br /> Clarity: SI2","Carat: 1.53 <br /> Price: 8996 <br /> Clarity: VS2","Carat: 1 <br /> Price: 4284 <br /> Clarity: SI2","Carat: 0.38 <br /> Price: 898 <br /> Clarity: VS2","Carat: 1 <br /> Price: 5292 <br /> Clarity: SI1","Carat: 1 <br /> Price: 2912 <br /> Clarity: SI1","Carat: 1.7 <br /> Price: 18055 <br /> Clarity: VS2","Carat: 0.55 <br /> Price: 2331 <br /> Clarity: VVS2","Carat: 1.13 <br /> Price: 7875 <br /> Clarity: VVS2","Carat: 0.57 <br /> Price: 1180 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 734 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 827 <br /> Clarity: VS1","Carat: 0.9 <br /> Price: 4712 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 645 <br /> Clarity: VVS2","Carat: 0.55 <br /> Price: 2079 <br /> Clarity: VS1","Carat: 0.91 <br /> Price: 3924 <br /> Clarity: VS2","Carat: 1.5 <br /> Price: 7098 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 801 <br /> Clarity: VVS2","Carat: 0.35 <br /> Price: 669 <br /> Clarity: SI2","Carat: 1.5 <br /> Price: 7955 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4242 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 838 <br /> Clarity: VVS1","Carat: 0.34 <br /> Price: 507 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 713 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 6416 <br /> Clarity: VS2","Carat: 1.04 <br /> Price: 4158 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 1153 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 2319 <br /> Clarity: VS2","Carat: 1.05 <br /> Price: 5134 <br /> Clarity: SI2","Carat: 1.07 <br /> Price: 4584 <br /> Clarity: SI2","Carat: 2.02 <br /> Price: 18398 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 745 <br /> Clarity: VS2","Carat: 0.73 <br /> Price: 3220 <br /> Clarity: VVS1","Carat: 0.71 <br /> Price: 1821 <br /> Clarity: VS2","Carat: 1.24 <br /> Price: 4858 <br /> Clarity: VS2","Carat: 0.26 <br /> Price: 635 <br /> Clarity: VVS1","Carat: 0.31 <br /> Price: 816 <br /> Clarity: VVS1","Carat: 0.38 <br /> Price: 800 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 689 <br /> Clarity: VS2","Carat: 1.2 <br /> Price: 4847 <br /> Clarity: SI2","Carat: 0.5 <br /> Price: 1257 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 1179 <br /> Clarity: VVS1","Carat: 1.75 <br /> Price: 8820 <br /> Clarity: SI2","Carat: 0.61 <br /> Price: 1827 <br /> Clarity: VVS2","Carat: 0.36 <br /> Price: 1215 <br /> Clarity: IF","Carat: 0.53 <br /> Price: 2407 <br /> Clarity: VVS2","Carat: 0.9 <br /> Price: 1599 <br /> Clarity: I1","Carat: 1.36 <br /> Price: 15247 <br /> Clarity: VVS1","Carat: 0.24 <br /> Price: 393 <br /> Clarity: VS1","Carat: 1.23 <br /> Price: 5504 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 488 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 6777 <br /> Clarity: VS1","Carat: 0.36 <br /> Price: 789 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 438 <br /> Clarity: VS2","Carat: 1.05 <br /> Price: 5480 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 926 <br /> Clarity: VVS1","Carat: 2.02 <br /> Price: 15231 <br /> Clarity: VS1","Carat: 1.21 <br /> Price: 6871 <br /> Clarity: VS1","Carat: 0.65 <br /> Price: 1219 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 1749 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 863 <br /> Clarity: VS2","Carat: 0.43 <br /> Price: 1118 <br /> Clarity: VVS1","Carat: 0.41 <br /> Price: 861 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 1075 <br /> Clarity: VVS2","Carat: 0.55 <br /> Price: 1165 <br /> Clarity: VS2","Carat: 0.52 <br /> Price: 1605 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 3496 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 5044 <br /> Clarity: SI1","Carat: 0.35 <br /> Price: 1035 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 1046 <br /> Clarity: VVS1","Carat: 1.03 <br /> Price: 6539 <br /> Clarity: VS1","Carat: 0.72 <br /> Price: 2591 <br /> Clarity: VVS2","Carat: 0.41 <br /> Price: 763 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 2270 <br /> Clarity: VS2","Carat: 0.47 <br /> Price: 784 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 734 <br /> Clarity: VS2","Carat: 2.01 <br /> Price: 17235 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1333 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 745 <br /> Clarity: VS2","Carat: 1.23 <br /> Price: 9122 <br /> Clarity: VS2","Carat: 1.14 <br /> Price: 9013 <br /> Clarity: VVS2","Carat: 1.53 <br /> Price: 7797 <br /> Clarity: SI1","Carat: 1.03 <br /> Price: 7752 <br /> Clarity: VS1","Carat: 1.71 <br /> Price: 8366 <br /> Clarity: SI2","Carat: 1.33 <br /> Price: 6480 <br /> Clarity: SI2","Carat: 1.04 <br /> Price: 5189 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 491 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 868 <br /> Clarity: IF","Carat: 1 <br /> Price: 4816 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 539 <br /> Clarity: VS1","Carat: 1.02 <br /> Price: 6797 <br /> Clarity: VVS2","Carat: 0.54 <br /> Price: 1240 <br /> Clarity: SI1","Carat: 0.83 <br /> Price: 3118 <br /> Clarity: SI1","Carat: 1.05 <br /> Price: 5557 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 872 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 730 <br /> Clarity: VVS2","Carat: 2.04 <br /> Price: 8257 <br /> Clarity: I1","Carat: 0.73 <br /> Price: 2371 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 6330 <br /> Clarity: VS2","Carat: 1 <br /> Price: 7260 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 652 <br /> Clarity: VS1","Carat: 0.44 <br /> Price: 1274 <br /> Clarity: VVS2","Carat: 0.53 <br /> Price: 1052 <br /> Clarity: SI1","Carat: 1.71 <br /> Price: 12338 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 6195 <br /> Clarity: SI1","Carat: 0.61 <br /> Price: 2324 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 2382 <br /> Clarity: VS2","Carat: 1.41 <br /> Price: 17216 <br /> Clarity: VVS2","Carat: 1.11 <br /> Price: 6737 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 1815 <br /> Clarity: VS2","Carat: 0.74 <br /> Price: 3353 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 4508 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 418 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 828 <br /> Clarity: VS2","Carat: 1 <br /> Price: 5219 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 2657 <br /> Clarity: VS2","Carat: 1 <br /> Price: 4697 <br /> Clarity: SI1","Carat: 0.72 <br /> Price: 2688 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 900 <br /> Clarity: VVS1","Carat: 1.59 <br /> Price: 11119 <br /> Clarity: VS1","Carat: 0.83 <br /> Price: 3010 <br /> Clarity: SI1","Carat: 1.61 <br /> Price: 9961 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 545 <br /> Clarity: VS1","Carat: 1.26 <br /> Price: 5792 <br /> Clarity: VS2","Carat: 0.56 <br /> Price: 1394 <br /> Clarity: SI1","Carat: 0.38 <br /> Price: 617 <br /> Clarity: VS2","Carat: 1.12 <br /> Price: 4290 <br /> Clarity: SI1","Carat: 0.74 <br /> Price: 3694 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 593 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 1235 <br /> Clarity: VVS1","Carat: 0.83 <br /> Price: 3103 <br /> Clarity: SI2","Carat: 0.5 <br /> Price: 1845 <br /> Clarity: VS2","Carat: 0.81 <br /> Price: 2301 <br /> Clarity: SI2","Carat: 0.71 <br /> Price: 4030 <br /> Clarity: VVS2","Carat: 0.31 <br /> Price: 687 <br /> Clarity: VVS2","Carat: 0.7 <br /> Price: 3107 <br /> Clarity: VS1","Carat: 0.53 <br /> Price: 2658 <br /> Clarity: IF","Carat: 0.33 <br /> Price: 666 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 645 <br /> Clarity: VS1","Carat: 1.17 <br /> Price: 7076 <br /> Clarity: VS2","Carat: 1.12 <br /> Price: 5363 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1436 <br /> Clarity: VS2","Carat: 1.21 <br /> Price: 5346 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 668 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 540 <br /> Clarity: SI2","Carat: 0.73 <br /> Price: 2596 <br /> Clarity: VS1","Carat: 1.04 <br /> Price: 5163 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 612 <br /> Clarity: VVS1","Carat: 0.23 <br /> Price: 530 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 506 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 5443 <br /> Clarity: SI1","Carat: 1 <br /> Price: 3920 <br /> Clarity: SI2","Carat: 0.54 <br /> Price: 1786 <br /> Clarity: SI1","Carat: 1.05 <br /> Price: 7247 <br /> Clarity: VS1","Carat: 0.35 <br /> Price: 614 <br /> Clarity: VS1","Carat: 0.56 <br /> Price: 2866 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 461 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 842 <br /> Clarity: VVS2","Carat: 0.68 <br /> Price: 1918 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 1778 <br /> Clarity: VVS1","Carat: 1.52 <br /> Price: 11392 <br /> Clarity: VVS2","Carat: 0.89 <br /> Price: 3663 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 720 <br /> Clarity: VS2","Carat: 2.4 <br /> Price: 17955 <br /> Clarity: SI2","Carat: 0.82 <br /> Price: 2939 <br /> Clarity: VS2","Carat: 0.27 <br /> Price: 470 <br /> Clarity: VS2","Carat: 0.55 <br /> Price: 1777 <br /> Clarity: VS2","Carat: 1 <br /> Price: 6020 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 4039 <br /> Clarity: SI2","Carat: 0.6 <br /> Price: 2131 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 942 <br /> Clarity: VS2","Carat: 0.36 <br /> Price: 912 <br /> Clarity: IF","Carat: 1.53 <br /> Price: 13994 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 936 <br /> Clarity: VVS2","Carat: 1.54 <br /> Price: 15197 <br /> Clarity: VVS2","Carat: 0.72 <br /> Price: 1832 <br /> Clarity: VS1","Carat: 0.59 <br /> Price: 2551 <br /> Clarity: IF","Carat: 0.31 <br /> Price: 1046 <br /> Clarity: VVS2","Carat: 0.7 <br /> Price: 2095 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 1002 <br /> Clarity: VS2","Carat: 0.72 <br /> Price: 3478 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 2511 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 789 <br /> Clarity: VVS1","Carat: 1.13 <br /> Price: 9796 <br /> Clarity: VS2","Carat: 0.42 <br /> Price: 1179 <br /> Clarity: VVS1","Carat: 0.7 <br /> Price: 2176 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 639 <br /> Clarity: VS2","Carat: 2.06 <br /> Price: 11764 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2239 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 1114 <br /> Clarity: VVS1","Carat: 1.08 <br /> Price: 5439 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 5897 <br /> Clarity: VS2","Carat: 0.56 <br /> Price: 1662 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 720 <br /> Clarity: VS1","Carat: 1.5 <br /> Price: 8316 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 489 <br /> Clarity: VS1","Carat: 1.02 <br /> Price: 5312 <br /> Clarity: VS2","Carat: 2.06 <br /> Price: 18293 <br /> Clarity: SI2","Carat: 1.19 <br /> Price: 5825 <br /> Clarity: SI1","Carat: 2 <br /> Price: 17084 <br /> Clarity: VS2","Carat: 0.74 <br /> Price: 5841 <br /> Clarity: VVS2","Carat: 0.46 <br /> Price: 1299 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1624 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 863 <br /> Clarity: VVS1","Carat: 1.53 <br /> Price: 11525 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 3077 <br /> Clarity: VS2","Carat: 2.65 <br /> Price: 16314 <br /> Clarity: SI2","Carat: 1.29 <br /> Price: 6140 <br /> Clarity: SI1","Carat: 0.57 <br /> Price: 1949 <br /> Clarity: VS1","Carat: 2.54 <br /> Price: 16353 <br /> Clarity: SI2","Carat: 1.59 <br /> Price: 13631 <br /> Clarity: SI1","Carat: 0.67 <br /> Price: 1914 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 5301 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1175 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 756 <br /> Clarity: SI1","Carat: 1 <br /> Price: 5119 <br /> Clarity: SI1","Carat: 0.35 <br /> Price: 906 <br /> Clarity: VS2","Carat: 1.34 <br /> Price: 10771 <br /> Clarity: VS1","Carat: 0.72 <br /> Price: 2148 <br /> Clarity: SI1","Carat: 0.8 <br /> Price: 2760 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 904 <br /> Clarity: SI1","Carat: 2.01 <br /> Price: 16582 <br /> Clarity: SI1","Carat: 1.03 <br /> Price: 6091 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 9697 <br /> Clarity: SI1","Carat: 1 <br /> Price: 5880 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 864 <br /> Clarity: VVS2","Carat: 0.37 <br /> Price: 926 <br /> Clarity: IF","Carat: 1 <br /> Price: 5606 <br /> Clarity: VS2","Carat: 0.39 <br /> Price: 1232 <br /> Clarity: VVS2","Carat: 1.22 <br /> Price: 5739 <br /> Clarity: SI2","Carat: 0.56 <br /> Price: 2016 <br /> Clarity: VS1","Carat: 0.24 <br /> Price: 678 <br /> Clarity: VVS1","Carat: 0.7 <br /> Price: 2066 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 4565 <br /> Clarity: VVS2","Carat: 0.26 <br /> Price: 569 <br /> Clarity: VVS2","Carat: 0.32 <br /> Price: 524 <br /> Clarity: VS1","Carat: 0.9 <br /> Price: 4101 <br /> Clarity: SI2","Carat: 0.75 <br /> Price: 2680 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 523 <br /> Clarity: VS2","Carat: 2.01 <br /> Price: 17849 <br /> Clarity: SI1","Carat: 1.13 <br /> Price: 3603 <br /> Clarity: I1","Carat: 1.01 <br /> Price: 7870 <br /> Clarity: VS1","Carat: 0.55 <br /> Price: 2314 <br /> Clarity: IF","Carat: 0.31 <br /> Price: 914 <br /> Clarity: IF","Carat: 1.05 <br /> Price: 8114 <br /> Clarity: VS1","Carat: 0.76 <br /> Price: 2399 <br /> Clarity: SI2","Carat: 1 <br /> Price: 5005 <br /> Clarity: SI1","Carat: 1.11 <br /> Price: 9377 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 432 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2414 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 687 <br /> Clarity: VS2","Carat: 0.55 <br /> Price: 1857 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 912 <br /> Clarity: VVS1","Carat: 0.24 <br /> Price: 521 <br /> Clarity: VVS1","Carat: 1.5 <br /> Price: 9533 <br /> Clarity: VS1","Carat: 0.81 <br /> Price: 4284 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 734 <br /> Clarity: VS2","Carat: 0.94 <br /> Price: 4230 <br /> Clarity: SI2","Carat: 1.21 <br /> Price: 9419 <br /> Clarity: VS2","Carat: 1.2 <br /> Price: 9641 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 422 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2972 <br /> Clarity: VS1","Carat: 0.92 <br /> Price: 5183 <br /> Clarity: VVS2","Carat: 2.35 <br /> Price: 17294 <br /> Clarity: SI2","Carat: 1.79 <br /> Price: 18419 <br /> Clarity: VS1","Carat: 2.01 <br /> Price: 14745 <br /> Clarity: SI2","Carat: 0.61 <br /> Price: 2753 <br /> Clarity: VVS1","Carat: 0.34 <br /> Price: 765 <br /> Clarity: VS2","Carat: 1.15 <br /> Price: 7662 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1649 <br /> Clarity: VS1","Carat: 0.42 <br /> Price: 898 <br /> Clarity: SI1","Carat: 2.02 <br /> Price: 17882 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 4939 <br /> Clarity: I1","Carat: 0.83 <br /> Price: 3078 <br /> Clarity: SI2","Carat: 1.36 <br /> Price: 11946 <br /> Clarity: VS1","Carat: 1 <br /> Price: 4295 <br /> Clarity: SI2","Carat: 1 <br /> Price: 4077 <br /> Clarity: SI2","Carat: 1.56 <br /> Price: 6966 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 540 <br /> Clarity: SI2","Carat: 1.06 <br /> Price: 7408 <br /> Clarity: VS2","Carat: 2.06 <br /> Price: 17650 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 589 <br /> Clarity: SI1","Carat: 1.16 <br /> Price: 4222 <br /> Clarity: SI1","Carat: 0.72 <br /> Price: 2074 <br /> Clarity: SI1","Carat: 0.58 <br /> Price: 1239 <br /> Clarity: SI1","Carat: 0.23 <br /> Price: 530 <br /> Clarity: VVS2","Carat: 0.74 <br /> Price: 2261 <br /> Clarity: SI2","Carat: 0.51 <br /> Price: 1569 <br /> Clarity: SI1","Carat: 0.44 <br /> Price: 1334 <br /> Clarity: VVS2","Carat: 0.33 <br /> Price: 838 <br /> Clarity: VVS1","Carat: 1.38 <br /> Price: 15968 <br /> Clarity: VVS1","Carat: 2.02 <br /> Price: 17153 <br /> Clarity: VS1","Carat: 0.4 <br /> Price: 737 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 645 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 5378 <br /> Clarity: SI1","Carat: 0.59 <br /> Price: 1743 <br /> Clarity: SI1","Carat: 0.61 <br /> Price: 1588 <br /> Clarity: SI2","Carat: 1.19 <br /> Price: 6781 <br /> Clarity: VS2","Carat: 1.04 <br /> Price: 7655 <br /> Clarity: VS1","Carat: 0.23 <br /> Price: 505 <br /> Clarity: VVS2","Carat: 0.59 <br /> Price: 2078 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 4252 <br /> Clarity: SI2","Carat: 0.53 <br /> Price: 1367 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 2570 <br /> Clarity: SI1","Carat: 0.55 <br /> Price: 1057 <br /> Clarity: SI2","Carat: 0.44 <br /> Price: 1274 <br /> Clarity: VVS2","Carat: 0.39 <br /> Price: 834 <br /> Clarity: SI2","Carat: 1.34 <br /> Price: 5065 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 628 <br /> Clarity: VS2","Carat: 0.56 <br /> Price: 1131 <br /> Clarity: VS2","Carat: 1 <br /> Price: 4155 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 553 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 898 <br /> Clarity: SI2","Carat: 2.01 <br /> Price: 10184 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 2719 <br /> Clarity: SI1","Carat: 0.91 <br /> Price: 3299 <br /> Clarity: VS2","Carat: 1.59 <br /> Price: 7811 <br /> Clarity: VS2","Carat: 1.15 <br /> Price: 5038 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1237 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 17279 <br /> Clarity: VVS1","Carat: 2.1 <br /> Price: 15511 <br /> Clarity: SI2","Carat: 0.63 <br /> Price: 1235 <br /> Clarity: SI2","Carat: 1.51 <br /> Price: 10274 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 2426 <br /> Clarity: VVS2","Carat: 0.32 <br /> Price: 589 <br /> Clarity: VVS1","Carat: 1.08 <br /> Price: 4771 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4653 <br /> Clarity: SI1","Carat: 1.19 <br /> Price: 7466 <br /> Clarity: VS1","Carat: 0.52 <br /> Price: 1815 <br /> Clarity: VS2","Carat: 2.2 <br /> Price: 13512 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 608 <br /> Clarity: VS2","Carat: 1.28 <br /> Price: 12210 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 394 <br /> Clarity: VS1","Carat: 1.61 <br /> Price: 7844 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 421 <br /> Clarity: SI2","Carat: 1.52 <br /> Price: 12958 <br /> Clarity: VS2","Carat: 0.49 <br /> Price: 1198 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 5422 <br /> Clarity: SI1","Carat: 1.17 <br /> Price: 5743 <br /> Clarity: SI2","Carat: 0.52 <br /> Price: 1600 <br /> Clarity: SI1","Carat: 0.74 <br /> Price: 3226 <br /> Clarity: VS1","Carat: 0.91 <br /> Price: 3979 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2445 <br /> Clarity: SI1","Carat: 1 <br /> Price: 6989 <br /> Clarity: VS1","Carat: 0.4 <br /> Price: 1229 <br /> Clarity: IF","Carat: 0.39 <br /> Price: 1089 <br /> Clarity: VVS1","Carat: 0.51 <br /> Price: 1237 <br /> Clarity: SI1","Carat: 0.35 <br /> Price: 754 <br /> Clarity: VS1","Carat: 1 <br /> Price: 4844 <br /> Clarity: SI1","Carat: 0.55 <br /> Price: 2079 <br /> Clarity: VS1","Carat: 0.38 <br /> Price: 700 <br /> Clarity: VS2","Carat: 0.92 <br /> Price: 3787 <br /> Clarity: SI2","Carat: 1.2 <br /> Price: 7258 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 10993 <br /> Clarity: VVS1","Carat: 1.02 <br /> Price: 4558 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 1872 <br /> Clarity: VVS1","Carat: 0.4 <br /> Price: 1059 <br /> Clarity: VVS1","Carat: 1.14 <br /> Price: 6500 <br /> Clarity: VS2","Carat: 1.3 <br /> Price: 6140 <br /> Clarity: SI2","Carat: 1 <br /> Price: 4200 <br /> Clarity: SI2","Carat: 0.57 <br /> Price: 1613 <br /> Clarity: VS1","Carat: 1.16 <br /> Price: 4668 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 11586 <br /> Clarity: SI1","Carat: 1.03 <br /> Price: 4679 <br /> Clarity: SI1","Carat: 0.35 <br /> Price: 906 <br /> Clarity: VVS2","Carat: 0.43 <br /> Price: 774 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 4001 <br /> Clarity: SI2","Carat: 0.76 <br /> Price: 2601 <br /> Clarity: VS1","Carat: 0.51 <br /> Price: 1431 <br /> Clarity: SI1","Carat: 0.55 <br /> Price: 1682 <br /> Clarity: VS1","Carat: 0.36 <br /> Price: 789 <br /> Clarity: VS2","Carat: 0.91 <br /> Price: 7131 <br /> Clarity: VVS2","Carat: 0.32 <br /> Price: 730 <br /> Clarity: VVS2","Carat: 0.71 <br /> Price: 1947 <br /> Clarity: SI2","Carat: 0.26 <br /> Price: 448 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 552 <br /> Clarity: SI1","Carat: 0.26 <br /> Price: 384 <br /> Clarity: SI1","Carat: 1.2 <br /> Price: 5718 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 550 <br /> Clarity: SI1","Carat: 3.02 <br /> Price: 10577 <br /> Clarity: I1","Carat: 1.27 <br /> Price: 5308 <br /> Clarity: SI2","Carat: 0.73 <br /> Price: 2865 <br /> Clarity: SI1","Carat: 1.52 <br /> Price: 8110 <br /> Clarity: VS1","Carat: 1.06 <br /> Price: 4507 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 898 <br /> Clarity: IF","Carat: 1.6 <br /> Price: 10824 <br /> Clarity: VS1","Carat: 0.6 <br /> Price: 2037 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 6499 <br /> Clarity: VS1","Carat: 2.51 <br /> Price: 15324 <br /> Clarity: SI1","Carat: 0.35 <br /> Price: 767 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 915 <br /> Clarity: VVS2","Carat: 0.33 <br /> Price: 738 <br /> Clarity: VS1","Carat: 1.09 <br /> Price: 4578 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 6295 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 5198 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 3774 <br /> Clarity: SI1","Carat: 0.57 <br /> Price: 1592 <br /> Clarity: IF","Carat: 0.4 <br /> Price: 596 <br /> Clarity: SI1","Carat: 1.17 <br /> Price: 9289 <br /> Clarity: VVS2","Carat: 0.34 <br /> Price: 477 <br /> Clarity: SI2","Carat: 1.53 <br /> Price: 11078 <br /> Clarity: SI2","Carat: 2.07 <br /> Price: 16957 <br /> Clarity: SI1","Carat: 0.36 <br /> Price: 933 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 593 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2765 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 756 <br /> Clarity: SI1","Carat: 0.54 <br /> Price: 1754 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 3799 <br /> Clarity: VVS2","Carat: 2.01 <br /> Price: 17987 <br /> Clarity: VS2","Carat: 0.59 <br /> Price: 2933 <br /> Clarity: VVS2","Carat: 0.54 <br /> Price: 1637 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 900 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 5067 <br /> Clarity: VVS2","Carat: 0.24 <br /> Price: 373 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 1002 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 867 <br /> Clarity: VVS1","Carat: 0.58 <br /> Price: 1600 <br /> Clarity: VS2","Carat: 0.6 <br /> Price: 2306 <br /> Clarity: VS1","Carat: 0.33 <br /> Price: 781 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 3061 <br /> Clarity: VS1","Carat: 0.46 <br /> Price: 904 <br /> Clarity: SI1","Carat: 0.88 <br /> Price: 3557 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4588 <br /> Clarity: SI2","Carat: 1.02 <br /> Price: 8627 <br /> Clarity: VVS2","Carat: 1.02 <br /> Price: 4946 <br /> Clarity: SI2","Carat: 0.56 <br /> Price: 1814 <br /> Clarity: SI1","Carat: 1.14 <br /> Price: 4173 <br /> Clarity: SI1","Carat: 0.97 <br /> Price: 3802 <br /> Clarity: SI2","Carat: 0.71 <br /> Price: 2130 <br /> Clarity: SI2","Carat: 0.55 <br /> Price: 1698 <br /> Clarity: SI1","Carat: 0.38 <br /> Price: 882 <br /> Clarity: VS1","Carat: 1.22 <br /> Price: 5233 <br /> Clarity: SI1","Carat: 0.29 <br /> Price: 504 <br /> Clarity: VS1","Carat: 0.26 <br /> Price: 679 <br /> Clarity: VVS1","Carat: 0.7 <br /> Price: 2717 <br /> Clarity: VS2","Carat: 0.42 <br /> Price: 1031 <br /> Clarity: VS1","Carat: 1.17 <br /> Price: 10115 <br /> Clarity: VS1","Carat: 1.29 <br /> Price: 6588 <br /> Clarity: SI1","Carat: 1.64 <br /> Price: 14201 <br /> Clarity: VS2","Carat: 0.73 <br /> Price: 2919 <br /> Clarity: VVS2","Carat: 0.41 <br /> Price: 1030 <br /> Clarity: VS1","Carat: 0.56 <br /> Price: 1723 <br /> Clarity: VS2","Carat: 0.44 <br /> Price: 795 <br /> Clarity: VS2","Carat: 0.77 <br /> Price: 3018 <br /> Clarity: VS2","Carat: 1.5 <br /> Price: 8492 <br /> Clarity: SI2","Carat: 1.51 <br /> Price: 7610 <br /> Clarity: SI2","Carat: 0.5 <br /> Price: 1923 <br /> Clarity: VS1","Carat: 1.69 <br /> Price: 13872 <br /> Clarity: SI1","Carat: 2.01 <br /> Price: 12182 <br /> Clarity: SI2","Carat: 1.21 <br /> Price: 7611 <br /> Clarity: VS2","Carat: 0.53 <br /> Price: 1393 <br /> Clarity: VS1","Carat: 1 <br /> Price: 5458 <br /> Clarity: SI1","Carat: 0.38 <br /> Price: 1257 <br /> Clarity: VVS2","Carat: 0.42 <br /> Price: 687 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2583 <br /> Clarity: SI2","Carat: 1.06 <br /> Price: 4749 <br /> Clarity: SI2","Carat: 0.75 <br /> Price: 2814 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 1822 <br /> Clarity: VS2","Carat: 1.59 <br /> Price: 12047 <br /> Clarity: VS1","Carat: 1.51 <br /> Price: 7162 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 776 <br /> Clarity: VS2","Carat: 0.35 <br /> Price: 1000 <br /> Clarity: VVS1","Carat: 0.4 <br /> Price: 1050 <br /> Clarity: VS2","Carat: 0.37 <br /> Price: 746 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 473 <br /> Clarity: VS1","Carat: 0.35 <br /> Price: 984 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 1257 <br /> Clarity: IF","Carat: 1 <br /> Price: 6285 <br /> Clarity: VS2","Carat: 2.01 <br /> Price: 15467 <br /> Clarity: SI1","Carat: 0.79 <br /> Price: 3256 <br /> Clarity: SI1","Carat: 0.73 <br /> Price: 2184 <br /> Clarity: SI1","Carat: 1.42 <br /> Price: 8529 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 2443 <br /> Clarity: VS2","Carat: 0.85 <br /> Price: 2278 <br /> Clarity: SI1","Carat: 1.26 <br /> Price: 8113 <br /> Clarity: VS2","Carat: 0.38 <br /> Price: 633 <br /> Clarity: VS2","Carat: 1.06 <br /> Price: 4530 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 1052 <br /> Clarity: IF","Carat: 0.54 <br /> Price: 1551 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 3462 <br /> Clarity: SI2","Carat: 1.21 <br /> Price: 4191 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 7942 <br /> Clarity: VVS2","Carat: 1.04 <br /> Price: 11279 <br /> Clarity: VVS1","Carat: 0.51 <br /> Price: 1662 <br /> Clarity: VS2","Carat: 0.42 <br /> Price: 663 <br /> Clarity: SI2","Carat: 1.83 <br /> Price: 6313 <br /> Clarity: I1","Carat: 0.51 <br /> Price: 1873 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 785 <br /> Clarity: VVS1","Carat: 0.42 <br /> Price: 948 <br /> Clarity: VS2","Carat: 1 <br /> Price: 7056 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 394 <br /> Clarity: SI2","Carat: 1.14 <br /> Price: 4789 <br /> Clarity: SI1","Carat: 0.72 <br /> Price: 3502 <br /> Clarity: VVS1","Carat: 0.28 <br /> Price: 470 <br /> Clarity: VS1","Carat: 1.35 <br /> Price: 7918 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 745 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 2610 <br /> Clarity: SI2","Carat: 0.9 <br /> Price: 3917 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 12236 <br /> Clarity: SI1","Carat: 0.53 <br /> Price: 1655 <br /> Clarity: VS1","Carat: 0.78 <br /> Price: 2365 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 462 <br /> Clarity: SI1","Carat: 1.13 <br /> Price: 10742 <br /> Clarity: VVS2","Carat: 0.81 <br /> Price: 3462 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 4562 <br /> Clarity: VS2","Carat: 0.59 <br /> Price: 2443 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 561 <br /> Clarity: VS1","Carat: 0.4 <br /> Price: 1050 <br /> Clarity: VS1","Carat: 0.33 <br /> Price: 965 <br /> Clarity: VVS1","Carat: 0.78 <br /> Price: 3035 <br /> Clarity: SI1","Carat: 1.55 <br /> Price: 10217 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 4466 <br /> Clarity: SI1","Carat: 0.72 <br /> Price: 2607 <br /> Clarity: VS2","Carat: 0.57 <br /> Price: 1308 <br /> Clarity: SI2","Carat: 0.34 <br /> Price: 447 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 11554 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 847 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 1353 <br /> Clarity: VVS2","Carat: 0.41 <br /> Price: 852 <br /> Clarity: SI1","Carat: 1.06 <br /> Price: 4774 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1098 <br /> Clarity: SI1","Carat: 0.77 <br /> Price: 3160 <br /> Clarity: SI1","Carat: 0.57 <br /> Price: 1606 <br /> Clarity: VS1","Carat: 0.8 <br /> Price: 2834 <br /> Clarity: VS2","Carat: 2.14 <br /> Price: 14138 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 878 <br /> Clarity: VVS1","Carat: 0.58 <br /> Price: 1671 <br /> Clarity: VS1","Carat: 1.5 <br /> Price: 7308 <br /> Clarity: VS2","Carat: 1.06 <br /> Price: 7679 <br /> Clarity: VS2","Carat: 0.43 <br /> Price: 850 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 7589 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 1621 <br /> Clarity: IF","Carat: 0.3 <br /> Price: 526 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 4191 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 12725 <br /> Clarity: IF","Carat: 0.61 <br /> Price: 2392 <br /> Clarity: VVS2","Carat: 1.52 <br /> Price: 6982 <br /> Clarity: VS2","Carat: 0.92 <br /> Price: 4155 <br /> Clarity: VS1","Carat: 1.52 <br /> Price: 10512 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 6669 <br /> Clarity: VS1","Carat: 1.02 <br /> Price: 3599 <br /> Clarity: SI2","Carat: 2.03 <br /> Price: 15272 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 1014 <br /> Clarity: VVS1","Carat: 2.22 <br /> Price: 6564 <br /> Clarity: I1","Carat: 0.75 <br /> Price: 2329 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 2549 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 1018 <br /> Clarity: IF","Carat: 0.7 <br /> Price: 2400 <br /> Clarity: VS2","Carat: 1.27 <br /> Price: 5518 <br /> Clarity: VS1","Carat: 0.44 <br /> Price: 1334 <br /> Clarity: VVS2","Carat: 0.36 <br /> Price: 663 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 590 <br /> Clarity: VVS2","Carat: 0.73 <br /> Price: 2841 <br /> Clarity: SI1","Carat: 1.1 <br /> Price: 4537 <br /> Clarity: SI2","Carat: 1.24 <br /> Price: 6438 <br /> Clarity: SI1","Carat: 0.35 <br /> Price: 984 <br /> Clarity: VS2","Carat: 0.44 <br /> Price: 1177 <br /> Clarity: IF","Carat: 0.43 <br /> Price: 848 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 605 <br /> Clarity: VVS2","Carat: 0.55 <br /> Price: 1668 <br /> Clarity: SI1","Carat: 0.36 <br /> Price: 600 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2066 <br /> Clarity: SI1","Carat: 1 <br /> Price: 5002 <br /> Clarity: VS2","Carat: 1.11 <br /> Price: 4196 <br /> Clarity: SI2","Carat: 0.61 <br /> Price: 2556 <br /> Clarity: VS2","Carat: 1.5 <br /> Price: 8167 <br /> Clarity: SI2","Carat: 1.24 <br /> Price: 4916 <br /> Clarity: SI2","Carat: 0.51 <br /> Price: 1644 <br /> Clarity: VS2","Carat: 1.16 <br /> Price: 7600 <br /> Clarity: VS2","Carat: 0.42 <br /> Price: 921 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 2711 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 868 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 814 <br /> Clarity: VVS1","Carat: 0.31 <br /> Price: 1046 <br /> Clarity: VVS2","Carat: 1.3 <br /> Price: 6188 <br /> Clarity: SI1","Carat: 0.55 <br /> Price: 2016 <br /> Clarity: VVS2","Carat: 0.72 <br /> Price: 2345 <br /> Clarity: SI2","Carat: 0.56 <br /> Price: 1694 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 810 <br /> Clarity: VS1","Carat: 0.52 <br /> Price: 2467 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 394 <br /> Clarity: SI2","Carat: 1.02 <br /> Price: 3884 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 945 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 2088 <br /> Clarity: SI1","Carat: 2.09 <br /> Price: 14024 <br /> Clarity: SI2","Carat: 1.2 <br /> Price: 8275 <br /> Clarity: VS1","Carat: 0.58 <br /> Price: 2529 <br /> Clarity: VVS2","Carat: 0.9 <br /> Price: 3890 <br /> Clarity: VS2","Carat: 2 <br /> Price: 18440 <br /> Clarity: SI1","Carat: 0.54 <br /> Price: 1745 <br /> Clarity: VS2","Carat: 1.2 <br /> Price: 5884 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 3811 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 754 <br /> Clarity: VVS2","Carat: 0.71 <br /> Price: 2209 <br /> Clarity: VS1","Carat: 0.34 <br /> Price: 995 <br /> Clarity: VVS1","Carat: 0.24 <br /> Price: 449 <br /> Clarity: VVS2","Carat: 0.52 <br /> Price: 1550 <br /> Clarity: VS2","Carat: 0.78 <br /> Price: 3601 <br /> Clarity: VS2","Carat: 1 <br /> Price: 3304 <br /> Clarity: SI2","Carat: 0.73 <br /> Price: 2527 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 16277 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 3342 <br /> Clarity: VS2","Carat: 1.7 <br /> Price: 10091 <br /> Clarity: SI2","Carat: 1.21 <br /> Price: 5140 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 4468 <br /> Clarity: SI2","Carat: 0.91 <br /> Price: 2676 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4563 <br /> Clarity: SI1","Carat: 0.83 <br /> Price: 3198 <br /> Clarity: SI1","Carat: 0.36 <br /> Price: 877 <br /> Clarity: IF","Carat: 0.3 <br /> Price: 684 <br /> Clarity: VVS2","Carat: 0.85 <br /> Price: 3183 <br /> Clarity: SI1","Carat: 0.23 <br /> Price: 484 <br /> Clarity: VVS1","Carat: 0.31 <br /> Price: 435 <br /> Clarity: SI2","Carat: 0.37 <br /> Price: 931 <br /> Clarity: VVS1","Carat: 1.31 <br /> Price: 4743 <br /> Clarity: SI2","Carat: 0.34 <br /> Price: 1133 <br /> Clarity: VVS1","Carat: 0.32 <br /> Price: 702 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 1053 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 3801 <br /> Clarity: SI2","Carat: 0.5 <br /> Price: 1935 <br /> Clarity: VVS2","Carat: 2.08 <br /> Price: 15065 <br /> Clarity: SI1","Carat: 0.36 <br /> Price: 608 <br /> Clarity: SI2","Carat: 0.74 <br /> Price: 2815 <br /> Clarity: VS1","Carat: 2.01 <br /> Price: 13052 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 743 <br /> Clarity: VS1","Carat: 0.42 <br /> Price: 690 <br /> Clarity: SI1","Carat: 0.38 <br /> Price: 833 <br /> Clarity: VS1","Carat: 1.02 <br /> Price: 4162 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 873 <br /> Clarity: SI1","Carat: 2 <br /> Price: 12610 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 1666 <br /> Clarity: SI1","Carat: 0.81 <br /> Price: 3042 <br /> Clarity: VS1","Carat: 0.33 <br /> Price: 579 <br /> Clarity: VS1","Carat: 0.76 <br /> Price: 2469 <br /> Clarity: SI2","Carat: 0.34 <br /> Price: 689 <br /> Clarity: VS2","Carat: 1.5 <br /> Price: 17203 <br /> Clarity: VVS2","Carat: 1.5 <br /> Price: 14386 <br /> Clarity: VS2","Carat: 2.1 <br /> Price: 13753 <br /> Clarity: VS2","Carat: 0.92 <br /> Price: 4791 <br /> Clarity: SI1","Carat: 1.24 <br /> Price: 7351 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 6470 <br /> Clarity: VVS2","Carat: 0.92 <br /> Price: 3941 <br /> Clarity: SI2","Carat: 1.46 <br /> Price: 5421 <br /> Clarity: SI2","Carat: 0.5 <br /> Price: 1173 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 2352 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 743 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 459 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 1408 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 5759 <br /> Clarity: VS2","Carat: 1.59 <br /> Price: 15843 <br /> Clarity: VVS2","Carat: 0.55 <br /> Price: 1179 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 872 <br /> Clarity: VS2","Carat: 0.92 <br /> Price: 4183 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 706 <br /> Clarity: VVS2","Carat: 0.71 <br /> Price: 2104 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 662 <br /> Clarity: VS2","Carat: 0.39 <br /> Price: 886 <br /> Clarity: SI1","Carat: 1.18 <br /> Price: 3965 <br /> Clarity: I1","Carat: 1.12 <br /> Price: 9461 <br /> Clarity: VVS2","Carat: 1.21 <br /> Price: 5672 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 1142 <br /> Clarity: VVS2","Carat: 2.01 <br /> Price: 10389 <br /> Clarity: SI1","Carat: 1.52 <br /> Price: 8151 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 800 <br /> Clarity: VS1","Carat: 0.93 <br /> Price: 2647 <br /> Clarity: I1","Carat: 0.28 <br /> Price: 487 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4189 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 1002 <br /> Clarity: VS2","Carat: 0.38 <br /> Price: 1026 <br /> Clarity: VS2","Carat: 2 <br /> Price: 17534 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 463 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 1333 <br /> Clarity: IF","Carat: 1.01 <br /> Price: 4564 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 5544 <br /> Clarity: SI1","Carat: 0.74 <br /> Price: 2877 <br /> Clarity: VS2","Carat: 0.37 <br /> Price: 531 <br /> Clarity: SI1","Carat: 1.53 <br /> Price: 12621 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 1236 <br /> Clarity: VVS1","Carat: 0.37 <br /> Price: 1041 <br /> Clarity: VS2","Carat: 0.56 <br /> Price: 2442 <br /> Clarity: IF","Carat: 0.4 <br /> Price: 722 <br /> Clarity: VS2","Carat: 1.32 <br /> Price: 9572 <br /> Clarity: VVS1","Carat: 0.5 <br /> Price: 1155 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 851 <br /> Clarity: VS2","Carat: 0.81 <br /> Price: 2901 <br /> Clarity: SI2","Carat: 0.9 <br /> Price: 3884 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 4712 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 5599 <br /> Clarity: SI1","Carat: 1.6 <br /> Price: 8808 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 521 <br /> Clarity: VS2","Carat: 1.2 <br /> Price: 6303 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2815 <br /> Clarity: SI1","Carat: 0.78 <br /> Price: 2781 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 473 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 988 <br /> Clarity: VVS1","Carat: 0.32 <br /> Price: 842 <br /> Clarity: IF","Carat: 0.53 <br /> Price: 1291 <br /> Clarity: SI1","Carat: 0.38 <br /> Price: 660 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 5759 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 936 <br /> Clarity: VVS2","Carat: 1.55 <br /> Price: 11957 <br /> Clarity: SI1","Carat: 0.73 <br /> Price: 2623 <br /> Clarity: VVS1","Carat: 1.51 <br /> Price: 7637 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 10023 <br /> Clarity: VS2","Carat: 1.03 <br /> Price: 5804 <br /> Clarity: SI1","Carat: 1.03 <br /> Price: 6662 <br /> Clarity: VS1","Carat: 0.53 <br /> Price: 1205 <br /> Clarity: SI1","Carat: 0.72 <br /> Price: 3012 <br /> Clarity: VS2","Carat: 0.27 <br /> Price: 470 <br /> Clarity: VS1","Carat: 0.67 <br /> Price: 1867 <br /> Clarity: VS2","Carat: 0.34 <br /> Price: 507 <br /> Clarity: SI2","Carat: 0.57 <br /> Price: 1613 <br /> Clarity: SI1","Carat: 0.23 <br /> Price: 505 <br /> Clarity: VVS2","Carat: 1.05 <br /> Price: 2705 <br /> Clarity: I1","Carat: 0.33 <br /> Price: 1040 <br /> Clarity: VVS2","Carat: 0.71 <br /> Price: 2137 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 1004 <br /> Clarity: IF","Carat: 1.01 <br /> Price: 4538 <br /> Clarity: SI1","Carat: 0.74 <br /> Price: 2805 <br /> Clarity: VS2","Carat: 0.52 <br /> Price: 2153 <br /> Clarity: VVS1","Carat: 1.13 <br /> Price: 4385 <br /> Clarity: SI1","Carat: 0.75 <br /> Price: 2801 <br /> Clarity: SI1","Carat: 1.42 <br /> Price: 9278 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 3437 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 793 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 6825 <br /> Clarity: VS1","Carat: 1.16 <br /> Price: 7589 <br /> Clarity: VS1","Carat: 0.71 <br /> Price: 2557 <br /> Clarity: VS1","Carat: 1.52 <br /> Price: 9552 <br /> Clarity: SI2","Carat: 0.51 <br /> Price: 1656 <br /> Clarity: VS2","Carat: 1.11 <br /> Price: 4096 <br /> Clarity: SI1","Carat: 2 <br /> Price: 15984 <br /> Clarity: SI2","Carat: 0.52 <br /> Price: 3284 <br /> Clarity: VVS2","Carat: 0.78 <br /> Price: 2469 <br /> Clarity: SI1","Carat: 1 <br /> Price: 4383 <br /> Clarity: SI2","Carat: 0.78 <br /> Price: 2826 <br /> Clarity: SI1","Carat: 0.75 <br /> Price: 3562 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 802 <br /> Clarity: VS1","Carat: 0.34 <br /> Price: 765 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 2838 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 950 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 574 <br /> Clarity: SI2","Carat: 0.9 <br /> Price: 4439 <br /> Clarity: SI1","Carat: 1.03 <br /> Price: 9245 <br /> Clarity: VVS2","Carat: 1.27 <br /> Price: 4779 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 521 <br /> Clarity: VS2","Carat: 1.2 <br /> Price: 5592 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 1243 <br /> Clarity: IF","Carat: 1.05 <br /> Price: 6196 <br /> Clarity: SI1","Carat: 0.6 <br /> Price: 1715 <br /> Clarity: VVS1","Carat: 0.48 <br /> Price: 1262 <br /> Clarity: VS1","Carat: 0.48 <br /> Price: 1270 <br /> Clarity: VS1","Carat: 1.07 <br /> Price: 4554 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 505 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 540 <br /> Clarity: SI1","Carat: 1.21 <br /> Price: 5707 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 3037 <br /> Clarity: VVS1","Carat: 1.12 <br /> Price: 5838 <br /> Clarity: SI1","Carat: 0.76 <br /> Price: 3075 <br /> Clarity: SI1","Carat: 1.28 <br /> Price: 6458 <br /> Clarity: SI2","Carat: 1.14 <br /> Price: 5819 <br /> Clarity: SI1","Carat: 1.74 <br /> Price: 13037 <br /> Clarity: VS1","Carat: 1.09 <br /> Price: 4078 <br /> Clarity: SI2","Carat: 1.82 <br /> Price: 13813 <br /> Clarity: VS1","Carat: 1.13 <br /> Price: 6980 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 773 <br /> Clarity: VS2","Carat: 0.51 <br /> Price: 1750 <br /> Clarity: VVS2","Carat: 0.5 <br /> Price: 1628 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 3618 <br /> Clarity: VVS2","Carat: 0.27 <br /> Price: 480 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 3871 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 515 <br /> Clarity: VVS2","Carat: 0.7 <br /> Price: 1817 <br /> Clarity: SI2","Carat: 1.08 <br /> Price: 5171 <br /> Clarity: VVS2","Carat: 1.22 <br /> Price: 6062 <br /> Clarity: SI1","Carat: 1.13 <br /> Price: 4113 <br /> Clarity: SI2","Carat: 0.51 <br /> Price: 1060 <br /> Clarity: VS2","Carat: 1.25 <br /> Price: 8281 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 776 <br /> Clarity: VVS2","Carat: 0.47 <br /> Price: 1421 <br /> Clarity: VVS1","Carat: 0.72 <br /> Price: 2295 <br /> Clarity: SI2","Carat: 0.72 <br /> Price: 2819 <br /> Clarity: VS1","Carat: 0.4 <br /> Price: 737 <br /> Clarity: SI1","Carat: 2.02 <br /> Price: 13502 <br /> Clarity: SI1","Carat: 0.43 <br /> Price: 1129 <br /> Clarity: VS1","Carat: 0.36 <br /> Price: 726 <br /> Clarity: VS1","Carat: 1.06 <br /> Price: 4719 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 3063 <br /> Clarity: VS1","Carat: 0.74 <br /> Price: 4013 <br /> Clarity: VVS1","Carat: 0.51 <br /> Price: 2035 <br /> Clarity: VVS2","Carat: 1.06 <br /> Price: 6411 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1323 <br /> Clarity: VS2","Carat: 1.51 <br /> Price: 9694 <br /> Clarity: SI1","Carat: 0.35 <br /> Price: 647 <br /> Clarity: SI1","Carat: 0.91 <br /> Price: 3991 <br /> Clarity: SI1","Carat: 0.53 <br /> Price: 1227 <br /> Clarity: SI2","Carat: 0.52 <br /> Price: 1689 <br /> Clarity: VS1","Carat: 0.25 <br /> Price: 633 <br /> Clarity: IF","Carat: 0.59 <br /> Price: 2018 <br /> Clarity: VS1","Carat: 0.72 <br /> Price: 1676 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2039 <br /> Clarity: SI2","Carat: 0.35 <br /> Price: 984 <br /> Clarity: VS2","Carat: 1.08 <br /> Price: 4627 <br /> Clarity: SI2","Carat: 0.79 <br /> Price: 2958 <br /> Clarity: VS1","Carat: 0.5 <br /> Price: 1691 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 4435 <br /> Clarity: SI1","Carat: 0.26 <br /> Price: 600 <br /> Clarity: VVS1","Carat: 0.73 <br /> Price: 2269 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 786 <br /> Clarity: VVS1","Carat: 0.72 <br /> Price: 2646 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 544 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 949 <br /> Clarity: VS2","Carat: 0.72 <br /> Price: 2724 <br /> Clarity: VS1","Carat: 2.01 <br /> Price: 11096 <br /> Clarity: SI2","Carat: 0.75 <br /> Price: 2372 <br /> Clarity: SI1","Carat: 1.53 <br /> Price: 9877 <br /> Clarity: VS1","Carat: 0.33 <br /> Price: 984 <br /> Clarity: VVS1","Carat: 1.01 <br /> Price: 4853 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2657 <br /> Clarity: VS2","Carat: 0.51 <br /> Price: 1882 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 4092 <br /> Clarity: SI1","Carat: 1.12 <br /> Price: 4498 <br /> Clarity: SI2","Carat: 0.94 <br /> Price: 3624 <br /> Clarity: SI2","Carat: 0.71 <br /> Price: 2036 <br /> Clarity: SI2","Carat: 0.54 <br /> Price: 1847 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 778 <br /> Clarity: VVS2","Carat: 2.28 <br /> Price: 16041 <br /> Clarity: SI2","Carat: 1.04 <br /> Price: 11382 <br /> Clarity: VVS1","Carat: 0.31 <br /> Price: 777 <br /> Clarity: VVS2","Carat: 0.5 <br /> Price: 1848 <br /> Clarity: SI1","Carat: 1 <br /> Price: 5330 <br /> Clarity: SI1","Carat: 1 <br /> Price: 8138 <br /> Clarity: VS2","Carat: 0.74 <br /> Price: 3133 <br /> Clarity: VS1","Carat: 1.02 <br /> Price: 3996 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 1018 <br /> Clarity: VS1","Carat: 1.78 <br /> Price: 12981 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 1168 <br /> Clarity: VVS1","Carat: 0.73 <br /> Price: 3370 <br /> Clarity: VS1","Carat: 1.07 <br /> Price: 5031 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 945 <br /> Clarity: VVS1","Carat: 0.43 <br /> Price: 943 <br /> Clarity: VVS2","Carat: 0.71 <br /> Price: 2441 <br /> Clarity: SI1","Carat: 0.59 <br /> Price: 1247 <br /> Clarity: SI2","Carat: 0.74 <br /> Price: 3922 <br /> Clarity: VVS2","Carat: 0.71 <br /> Price: 2218 <br /> Clarity: SI1","Carat: 1 <br /> Price: 4081 <br /> Clarity: SI2","Carat: 1.02 <br /> Price: 8303 <br /> Clarity: VS1","Carat: 0.51 <br /> Price: 2082 <br /> Clarity: VVS2","Carat: 0.43 <br /> Price: 921 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 561 <br /> Clarity: VS2","Carat: 1.58 <br /> Price: 10738 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 7929 <br /> Clarity: SI2","Carat: 1.52 <br /> Price: 11105 <br /> Clarity: VS2","Carat: 0.41 <br /> Price: 889 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 3360 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 984 <br /> Clarity: IF","Carat: 1.13 <br /> Price: 9525 <br /> Clarity: IF","Carat: 1.29 <br /> Price: 6509 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 1184 <br /> Clarity: VVS2","Carat: 0.76 <br /> Price: 2517 <br /> Clarity: VS2","Carat: 0.43 <br /> Price: 669 <br /> Clarity: SI2","Carat: 0.71 <br /> Price: 3006 <br /> Clarity: VS1","Carat: 0.34 <br /> Price: 477 <br /> Clarity: SI2","Carat: 0.26 <br /> Price: 532 <br /> Clarity: VVS1","Carat: 0.31 <br /> Price: 544 <br /> Clarity: SI1","Carat: 0.35 <br /> Price: 1254 <br /> Clarity: IF","Carat: 0.4 <br /> Price: 1250 <br /> Clarity: VS1","Carat: 0.27 <br /> Price: 682 <br /> Clarity: VVS2","Carat: 0.33 <br /> Price: 868 <br /> Clarity: IF","Carat: 1.62 <br /> Price: 9526 <br /> Clarity: VS1","Carat: 1.05 <br /> Price: 10915 <br /> Clarity: VVS1","Carat: 1.01 <br /> Price: 8119 <br /> Clarity: VS2","Carat: 0.96 <br /> Price: 1808 <br /> Clarity: I1","Carat: 0.33 <br /> Price: 743 <br /> Clarity: VS2","Carat: 1.73 <br /> Price: 8220 <br /> Clarity: I1","Carat: 1.21 <br /> Price: 5556 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 1810 <br /> Clarity: VVS1","Carat: 1.29 <br /> Price: 4277 <br /> Clarity: I1","Carat: 1.24 <br /> Price: 10388 <br /> Clarity: VS1","Carat: 0.8 <br /> Price: 3091 <br /> Clarity: SI1","Carat: 2.01 <br /> Price: 14792 <br /> Clarity: VS1","Carat: 0.76 <br /> Price: 4223 <br /> Clarity: VVS2","Carat: 0.74 <br /> Price: 2447 <br /> Clarity: SI2","Carat: 1 <br /> Price: 5639 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 1881 <br /> Clarity: VVS1","Carat: 0.71 <br /> Price: 2386 <br /> Clarity: SI1","Carat: 1.09 <br /> Price: 4682 <br /> Clarity: SI2","Carat: 1.37 <br /> Price: 10226 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4497 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 949 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 4129 <br /> Clarity: SI1","Carat: 2.03 <br /> Price: 15095 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 544 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 3730 <br /> Clarity: SI1","Carat: 0.36 <br /> Price: 505 <br /> Clarity: SI2","Carat: 0.37 <br /> Price: 833 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 828 <br /> Clarity: VS2","Carat: 1 <br /> Price: 3265 <br /> Clarity: VS2","Carat: 0.77 <br /> Price: 3121 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 564 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 702 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 936 <br /> Clarity: VVS1","Carat: 1.09 <br /> Price: 5164 <br /> Clarity: SI2","Carat: 1.05 <br /> Price: 4374 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 710 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 878 <br /> Clarity: VVS1","Carat: 0.72 <br /> Price: 2795 <br /> Clarity: VS1","Carat: 0.8 <br /> Price: 2451 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2230 <br /> Clarity: SI1","Carat: 1.05 <br /> Price: 2293 <br /> Clarity: I1","Carat: 0.32 <br /> Price: 612 <br /> Clarity: SI1","Carat: 0.93 <br /> Price: 4666 <br /> Clarity: SI1","Carat: 0.73 <br /> Price: 3567 <br /> Clarity: VVS1","Carat: 1.01 <br /> Price: 7258 <br /> Clarity: VS2","Carat: 0.75 <br /> Price: 1958 <br /> Clarity: SI1","Carat: 0.72 <br /> Price: 3035 <br /> Clarity: VS1","Carat: 0.76 <br /> Price: 2329 <br /> Clarity: SI2","Carat: 2.01 <br /> Price: 15955 <br /> Clarity: SI2","Carat: 1.02 <br /> Price: 5038 <br /> Clarity: SI1","Carat: 2.06 <br /> Price: 14803 <br /> Clarity: SI2","Carat: 0.51 <br /> Price: 2005 <br /> Clarity: VVS1","Carat: 0.35 <br /> Price: 551 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 684 <br /> Clarity: VVS2","Carat: 0.39 <br /> Price: 1024 <br /> Clarity: VS1","Carat: 0.35 <br /> Price: 952 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4149 <br /> Clarity: SI2","Carat: 0.52 <br /> Price: 1266 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 990 <br /> Clarity: VVS2","Carat: 0.55 <br /> Price: 1676 <br /> Clarity: VS2","Carat: 0.24 <br /> Price: 419 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 4203 <br /> Clarity: VS2","Carat: 0.41 <br /> Price: 625 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 868 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 602 <br /> Clarity: VVS2","Carat: 0.5 <br /> Price: 1935 <br /> Clarity: VVS2","Carat: 1 <br /> Price: 7848 <br /> Clarity: VVS1","Carat: 1.23 <br /> Price: 7577 <br /> Clarity: VS1","Carat: 1.3 <br /> Price: 6928 <br /> Clarity: VS1","Carat: 0.5 <br /> Price: 1838 <br /> Clarity: VVS1","Carat: 0.7 <br /> Price: 2877 <br /> Clarity: VS1","Carat: 0.61 <br /> Price: 1829 <br /> Clarity: SI1","Carat: 0.28 <br /> Price: 481 <br /> Clarity: SI1","Carat: 0.92 <br /> Price: 3057 <br /> Clarity: SI2","Carat: 0.59 <br /> Price: 1445 <br /> Clarity: SI1","Carat: 0.27 <br /> Price: 620 <br /> Clarity: VVS1","Carat: 1 <br /> Price: 3965 <br /> Clarity: SI2","Carat: 1.52 <br /> Price: 13903 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 1662 <br /> Clarity: SI2","Carat: 0.59 <br /> Price: 3791 <br /> Clarity: VVS1","Carat: 2.04 <br /> Price: 16874 <br /> Clarity: VS2","Carat: 0.52 <br /> Price: 1875 <br /> Clarity: VS1","Carat: 1.08 <br /> Price: 5154 <br /> Clarity: SI1","Carat: 0.34 <br /> Price: 1071 <br /> Clarity: VVS1","Carat: 0.5 <br /> Price: 1374 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 946 <br /> Clarity: IF","Carat: 0.71 <br /> Price: 3247 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 758 <br /> Clarity: VS2","Carat: 1.77 <br /> Price: 10051 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 658 <br /> Clarity: VS2","Carat: 1.01 <br /> Price: 6516 <br /> Clarity: VS2","Carat: 1.53 <br /> Price: 11434 <br /> Clarity: VS2","Carat: 1.7 <br /> Price: 10337 <br /> Clarity: VVS2","Carat: 0.53 <br /> Price: 1797 <br /> Clarity: VVS2","Carat: 1.15 <br /> Price: 8349 <br /> Clarity: VVS2","Carat: 1.01 <br /> Price: 4751 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 949 <br /> Clarity: VVS2","Carat: 0.41 <br /> Price: 755 <br /> Clarity: SI1","Carat: 0.82 <br /> Price: 3781 <br /> Clarity: VVS2","Carat: 0.53 <br /> Price: 1727 <br /> Clarity: VS2","Carat: 0.26 <br /> Price: 517 <br /> Clarity: VVS2","Carat: 0.7 <br /> Price: 2237 <br /> Clarity: SI2","Carat: 1 <br /> Price: 4077 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 4241 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 1356 <br /> Clarity: VVS2","Carat: 1.05 <br /> Price: 4679 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 2344 <br /> Clarity: I1","Carat: 1.01 <br /> Price: 4688 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 4619 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 552 <br /> Clarity: SI1","Carat: 0.73 <br /> Price: 2579 <br /> Clarity: SI1","Carat: 0.39 <br /> Price: 790 <br /> Clarity: VS1","Carat: 1.19 <br /> Price: 6857 <br /> Clarity: SI1","Carat: 0.55 <br /> Price: 1916 <br /> Clarity: VVS2","Carat: 0.38 <br /> Price: 766 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 2428 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 2517 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 515 <br /> Clarity: VVS2","Carat: 1.07 <br /> Price: 3775 <br /> Clarity: SI2","Carat: 0.63 <br /> Price: 2313 <br /> Clarity: SI1","Carat: 1.5 <br /> Price: 12787 <br /> Clarity: VS2","Carat: 0.6 <br /> Price: 1433 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 477 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 886 <br /> Clarity: IF","Carat: 0.43 <br /> Price: 713 <br /> Clarity: I1","Carat: 0.25 <br /> Price: 563 <br /> Clarity: VS1","Carat: 1.5 <br /> Price: 12862 <br /> Clarity: VS1","Carat: 1.36 <br /> Price: 14236 <br /> Clarity: VVS2","Carat: 1.21 <br /> Price: 5387 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 1122 <br /> Clarity: IF","Carat: 0.34 <br /> Price: 689 <br /> Clarity: VS2","Carat: 0.43 <br /> Price: 1264 <br /> Clarity: VVS1","Carat: 0.98 <br /> Price: 3731 <br /> Clarity: SI2","Carat: 1.09 <br /> Price: 5042 <br /> Clarity: SI2","Carat: 1.22 <br /> Price: 8067 <br /> Clarity: VS2","Carat: 0.81 <br /> Price: 2095 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 780 <br /> Clarity: SI1","Carat: 1.04 <br /> Price: 9281 <br /> Clarity: VVS2","Carat: 0.9 <br /> Price: 3595 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 506 <br /> Clarity: SI2","Carat: 2.06 <br /> Price: 14488 <br /> Clarity: SI1","Carat: 1.2 <br /> Price: 8596 <br /> Clarity: VS1","Carat: 0.57 <br /> Price: 2360 <br /> Clarity: VS1","Carat: 0.34 <br /> Price: 596 <br /> Clarity: VS2","Carat: 0.96 <br /> Price: 3312 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 707 <br /> Clarity: VVS2","Carat: 1.7 <br /> Price: 12874 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 925 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 2400 <br /> Clarity: VS2","Carat: 0.63 <br /> Price: 2481 <br /> Clarity: VVS2","Carat: 0.46 <br /> Price: 1172 <br /> Clarity: VS2","Carat: 1.1 <br /> Price: 5267 <br /> Clarity: SI1","Carat: 1.54 <br /> Price: 4980 <br /> Clarity: I1","Carat: 1.52 <br /> Price: 9952 <br /> Clarity: SI1","Carat: 0.52 <br /> Price: 1577 <br /> Clarity: SI1","Carat: 0.51 <br /> Price: 1781 <br /> Clarity: VS1","Carat: 1.31 <br /> Price: 9797 <br /> Clarity: VS1","Carat: 0.38 <br /> Price: 700 <br /> Clarity: VS1","Carat: 0.42 <br /> Price: 992 <br /> Clarity: SI1","Carat: 0.91 <br /> Price: 3506 <br /> Clarity: SI1","Carat: 1 <br /> Price: 3889 <br /> Clarity: SI2","Carat: 0.44 <br /> Price: 990 <br /> Clarity: SI1","Carat: 0.8 <br /> Price: 2927 <br /> Clarity: SI1","Carat: 0.37 <br /> Price: 708 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 938 <br /> Clarity: IF","Carat: 1.04 <br /> Price: 4323 <br /> Clarity: SI2","Carat: 0.34 <br /> Price: 1030 <br /> Clarity: VVS1","Carat: 1.51 <br /> Price: 12947 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 684 <br /> Clarity: SI1","Carat: 1.71 <br /> Price: 10309 <br /> Clarity: VS1","Carat: 0.25 <br /> Price: 633 <br /> Clarity: VVS2","Carat: 1.16 <br /> Price: 4560 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 645 <br /> Clarity: VS1","Carat: 1.52 <br /> Price: 8521 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 4140 <br /> Clarity: SI2","Carat: 0.3 <br /> Price: 684 <br /> Clarity: VVS2","Carat: 0.34 <br /> Price: 805 <br /> Clarity: VS2","Carat: 1.31 <br /> Price: 13406 <br /> Clarity: VVS1","Carat: 1.21 <br /> Price: 5285 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 907 <br /> Clarity: VVS1","Carat: 0.36 <br /> Price: 729 <br /> Clarity: VS2","Carat: 0.33 <br /> Price: 925 <br /> Clarity: IF","Carat: 0.39 <br /> Price: 534 <br /> Clarity: SI1","Carat: 0.55 <br /> Price: 1698 <br /> Clarity: VS2","Carat: 0.43 <br /> Price: 716 <br /> Clarity: SI2","Carat: 0.32 <br /> Price: 612 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 1192 <br /> Clarity: VVS2","Carat: 1.03 <br /> Price: 5241 <br /> Clarity: SI1","Carat: 1.55 <br /> Price: 13912 <br /> Clarity: VS2","Carat: 0.35 <br /> Price: 767 <br /> Clarity: VS2","Carat: 1.5 <br /> Price: 7715 <br /> Clarity: SI2","Carat: 1.09 <br /> Price: 7238 <br /> Clarity: VS2","Carat: 0.57 <br /> Price: 1819 <br /> Clarity: VS1","Carat: 0.5 <br /> Price: 1624 <br /> Clarity: VS2","Carat: 0.53 <br /> Price: 1957 <br /> Clarity: IF","Carat: 0.31 <br /> Price: 707 <br /> Clarity: VVS2","Carat: 0.32 <br /> Price: 730 <br /> Clarity: VVS2","Carat: 1.61 <br /> Price: 9439 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 4503 <br /> Clarity: VS2","Carat: 0.7 <br /> Price: 1808 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1281 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 633 <br /> Clarity: VVS2","Carat: 0.32 <br /> Price: 645 <br /> Clarity: VVS2","Carat: 0.56 <br /> Price: 1915 <br /> Clarity: VS1","Carat: 0.33 <br /> Price: 928 <br /> Clarity: VS2","Carat: 0.42 <br /> Price: 798 <br /> Clarity: VVS2","Carat: 0.38 <br /> Price: 1245 <br /> Clarity: VVS1","Carat: 0.76 <br /> Price: 2624 <br /> Clarity: SI2","Carat: 0.91 <br /> Price: 5484 <br /> Clarity: VS2","Carat: 1.11 <br /> Price: 4687 <br /> Clarity: VS1","Carat: 1 <br /> Price: 5557 <br /> Clarity: VS1","Carat: 0.51 <br /> Price: 1355 <br /> Clarity: SI1","Carat: 1.17 <br /> Price: 4826 <br /> Clarity: SI2","Carat: 1.06 <br /> Price: 9680 <br /> Clarity: VVS2","Carat: 0.33 <br /> Price: 723 <br /> Clarity: VS2","Carat: 0.58 <br /> Price: 1332 <br /> Clarity: SI1","Carat: 1.38 <br /> Price: 5969 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 692 <br /> Clarity: VS1","Carat: 1.2 <br /> Price: 4838 <br /> Clarity: SI1","Carat: 0.26 <br /> Price: 545 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 411 <br /> Clarity: VS1","Carat: 1.32 <br /> Price: 7480 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 911 <br /> Clarity: VS2","Carat: 0.23 <br /> Price: 538 <br /> Clarity: VVS2","Carat: 1 <br /> Price: 10577 <br /> Clarity: VVS2","Carat: 0.33 <br /> Price: 433 <br /> Clarity: SI2","Carat: 1.01 <br /> Price: 7127 <br /> Clarity: VS2","Carat: 0.54 <br /> Price: 1129 <br /> Clarity: VS2","Carat: 0.42 <br /> Price: 921 <br /> Clarity: VS1","Carat: 0.72 <br /> Price: 2718 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 3605 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 573 <br /> Clarity: SI1","Carat: 0.42 <br /> Price: 743 <br /> Clarity: SI1","Carat: 0.7 <br /> Price: 3618 <br /> Clarity: VS1","Carat: 0.51 <br /> Price: 1051 <br /> Clarity: SI2","Carat: 2.02 <br /> Price: 12267 <br /> Clarity: SI1","Carat: 0.9 <br /> Price: 4124 <br /> Clarity: VS2","Carat: 0.91 <br /> Price: 4123 <br /> Clarity: SI2","Carat: 1.22 <br /> Price: 6887 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 485 <br /> Clarity: SI2","Carat: 1.51 <br /> Price: 15303 <br /> Clarity: VVS2","Carat: 1.13 <br /> Price: 5012 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 702 <br /> Clarity: VVS1","Carat: 0.9 <br /> Price: 4921 <br /> Clarity: SI1","Carat: 1.01 <br /> Price: 6132 <br /> Clarity: SI1","Carat: 0.75 <br /> Price: 3462 <br /> Clarity: VS1","Carat: 1.2 <br /> Price: 8964 <br /> Clarity: VS2","Carat: 0.91 <br /> Price: 4514 <br /> Clarity: VS1","Carat: 0.7 <br /> Price: 1782 <br /> Clarity: VS1","Carat: 1.53 <br /> Price: 9896 <br /> Clarity: VS1","Carat: 1.02 <br /> Price: 2751 <br /> Clarity: I1","Carat: 0.52 <br /> Price: 1888 <br /> Clarity: VS2","Carat: 0.52 <br /> Price: 2301 <br /> Clarity: VVS2","Carat: 0.92 <br /> Price: 3763 <br /> Clarity: SI2","Carat: 3.01 <br /> Price: 16538 <br /> Clarity: I1","Carat: 0.33 <br /> Price: 891 <br /> Clarity: IF","Carat: 1.23 <br /> Price: 5412 <br /> Clarity: SI2","Carat: 0.26 <br /> Price: 427 <br /> Clarity: VVS2","Carat: 0.3 <br /> Price: 457 <br /> Clarity: VS1","Carat: 0.58 <br /> Price: 2494 <br /> Clarity: VVS2","Carat: 1.21 <br /> Price: 6125 <br /> Clarity: SI2","Carat: 0.31 <br /> Price: 707 <br /> Clarity: VVS1","Carat: 0.22 <br /> Price: 470 <br /> Clarity: SI1","Carat: 1.22 <br /> Price: 6156 <br /> Clarity: VS2","Carat: 1.58 <br /> Price: 12799 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 915 <br /> Clarity: VS2","Carat: 0.5 <br /> Price: 1415 <br /> Clarity: SI1","Carat: 1.2 <br /> Price: 11301 <br /> Clarity: VVS2","Carat: 0.41 <br /> Price: 1079 <br /> Clarity: VS1","Carat: 0.31 <br /> Price: 1046 <br /> Clarity: VVS1","Carat: 1.51 <br /> Price: 9140 <br /> Clarity: SI2","Carat: 0.51 <br /> Price: 1799 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 4504 <br /> Clarity: SI2","Carat: 0.37 <br /> Price: 649 <br /> Clarity: VS2","Carat: 0.4 <br /> Price: 711 <br /> Clarity: SI1","Carat: 1.51 <br /> Price: 14482 <br /> Clarity: VS2","Carat: 1.39 <br /> Price: 6694 <br /> Clarity: SI1","Carat: 1.11 <br /> Price: 3854 <br /> Clarity: SI2","Carat: 1.22 <br /> Price: 6012 <br /> Clarity: VS1","Carat: 0.35 <br /> Price: 839 <br /> Clarity: VS1","Carat: 0.27 <br /> Price: 622 <br /> Clarity: VVS1","Carat: 0.3 <br /> Price: 624 <br /> Clarity: VS1","Carat: 1 <br /> Price: 6377 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 4724 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 574 <br /> Clarity: SI2","Carat: 1 <br /> Price: 4082 <br /> Clarity: SI2","Carat: 1.2 <br /> Price: 5645 <br /> Clarity: SI2","Carat: 0.77 <br /> Price: 3350 <br /> Clarity: VS1","Carat: 0.35 <br /> Price: 923 <br /> Clarity: VS2","Carat: 1.09 <br /> Price: 5816 <br /> Clarity: SI1","Carat: 0.8 <br /> Price: 4268 <br /> Clarity: IF","Carat: 1.23 <br /> Price: 9230 <br /> Clarity: VS1","Carat: 1.01 <br /> Price: 7014 <br /> Clarity: VS2","Carat: 0.36 <br /> Price: 819 <br /> Clarity: VS1","Carat: 2.22 <br /> Price: 18363 <br /> Clarity: VS2","Carat: 0.75 <br /> Price: 1588 <br /> Clarity: SI2","Carat: 1.02 <br /> Price: 5169 <br /> Clarity: SI1","Carat: 0.93 <br /> Price: 3376 <br /> Clarity: SI2","Carat: 0.76 <br /> Price: 2287 <br /> Clarity: SI2","Carat: 0.33 <br /> Price: 743 <br /> Clarity: VS2","Carat: 0.57 <br /> Price: 2298 <br /> Clarity: VVS2","Carat: 0.31 <br /> Price: 766 <br /> Clarity: VVS2","Carat: 0.62 <br /> Price: 1415 <br /> Clarity: VVS2","Carat: 0.7 <br /> Price: 2401 <br /> Clarity: SI1","Carat: 0.5 <br /> Price: 1612 <br /> Clarity: VS2","Carat: 0.42 <br /> Price: 931 <br /> Clarity: SI1","Carat: 0.31 <br /> Price: 626 <br /> Clarity: VS2","Carat: 0.55 <br /> Price: 1410 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 1061 <br /> Clarity: VS2","Carat: 0.47 <br /> Price: 983 <br /> Clarity: SI1","Carat: 1.09 <br /> Price: 4090 <br /> Clarity: SI1","Carat: 0.33 <br /> Price: 579 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 1715 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 524 <br /> Clarity: VS2","Carat: 0.91 <br /> Price: 3210 <br /> Clarity: VS1","Carat: 0.9 <br /> Price: 4353 <br /> Clarity: VS1","Carat: 0.36 <br /> Price: 794 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 3332 <br /> Clarity: VVS1","Carat: 2.01 <br /> Price: 18172 <br /> Clarity: VS2","Carat: 1.51 <br /> Price: 12437 <br /> Clarity: VS2","Carat: 0.32 <br /> Price: 828 <br /> Clarity: VVS2","Carat: 0.55 <br /> Price: 1238 <br /> Clarity: VS1","Carat: 1.02 <br /> Price: 4398 <br /> Clarity: SI2","Carat: 0.7 <br /> Price: 1115 <br /> Clarity: I1","Carat: 1.52 <br /> Price: 9780 <br /> Clarity: SI1","Carat: 0.3 <br /> Price: 1013 <br /> Clarity: VVS2","Carat: 1.96 <br /> Price: 13099 <br /> Clarity: SI2","Carat: 0.39 <br /> Price: 581 <br /> Clarity: VS2","Carat: 1.32 <br /> Price: 4140 <br /> Clarity: I1","Carat: 0.71 <br /> Price: 2300 <br /> Clarity: VS2","Carat: 1.23 <br /> Price: 12848 <br /> Clarity: VVS2","Carat: 1 <br /> Price: 4321 <br /> Clarity: SI2","Carat: 1.3 <br /> Price: 6442 <br /> Clarity: VS2","Carat: 1.57 <br /> Price: 17175 <br /> Clarity: VS1","Carat: 1.25 <br /> Price: 5670 <br /> Clarity: SI2","Carat: 0.65 <br /> Price: 2009 <br /> Clarity: VS2","Carat: 0.41 <br /> Price: 949 <br /> Clarity: VS2","Carat: 1.02 <br /> Price: 7677 <br /> Clarity: VS2","Carat: 1.2 <br /> Price: 11530 <br /> Clarity: VVS2","Carat: 0.58 <br /> Price: 1424 <br /> Clarity: SI1","Carat: 0.57 <br /> Price: 1884 <br /> Clarity: VS1","Carat: 0.91 <br /> Price: 3546 <br /> Clarity: SI2","Carat: 1.09 <br /> Price: 7081 <br /> Clarity: VS2","Carat: 1.52 <br /> Price: 16492 <br /> Clarity: VVS1","Carat: 0.28 <br /> Price: 787 <br /> Clarity: IF","Carat: 0.81 <br /> Price: 4127 <br /> Clarity: VVS2","Carat: 2.02 <br /> Price: 18236 <br /> Clarity: SI2","Carat: 0.41 <br /> Price: 870 <br /> Clarity: SI1","Carat: 1.57 <br /> Price: 10093 <br /> Clarity: VS2","Carat: 0.74 <br /> Price: 1632 <br /> Clarity: SI2","Carat: 0.43 <br /> Price: 1358 <br /> Clarity: VVS1","Carat: 0.33 <br /> Price: 610 <br /> Clarity: VS1","Carat: 0.32 <br /> Price: 842 <br /> Clarity: VVS2","Carat: 0.74 <br /> Price: 3590 <br /> Clarity: VS1","Carat: 1.37 <br /> Price: 6337 <br /> Clarity: VS2","Carat: 0.58 <br /> Price: 1809 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 4484 <br /> Clarity: VS2","Carat: 0.9 <br /> Price: 3105 <br /> Clarity: SI2","Carat: 1.4 <br /> Price: 15134 <br /> Clarity: VVS1","Carat: 0.54 <br /> Price: 1680 <br /> Clarity: VS2","Carat: 0.92 <br /> Price: 3545 <br /> Clarity: SI1","Carat: 0.71 <br /> Price: 2015 <br /> Clarity: SI2","Carat: 1.22 <br /> Price: 8014 <br /> Clarity: VVS2","Carat: 1.2 <br /> Price: 5107 <br /> Clarity: SI1","Carat: 1.4 <br /> Price: 8216 <br /> Clarity: VS2","Carat: 0.71 <br /> Price: 2345 <br /> Clarity: SI1","Carat: 0.4 <br /> Price: 816 <br /> Clarity: VS2","Carat: 0.3 <br /> Price: 678 <br /> Clarity: VVS1","Carat: 1.31 <br /> Price: 5450 <br /> Clarity: SI1","Carat: 0.41 <br /> Price: 1076 <br /> Clarity: VS1","Carat: 0.41 <br /> Price: 1085 <br /> Clarity: VS2","Carat: 0.36 <br /> Price: 1013 <br /> Clarity: VS2","Carat: 1.06 <br /> Price: 13588 <br /> Clarity: VVS2","Carat: 0.56 <br /> Price: 957 <br /> Clarity: SI1","Carat: 1.02 <br /> Price: 3656 <br /> Clarity: SI2","Carat: 1 <br /> Price: 6377 <br /> Clarity: VS1","Carat: 0.77 <br /> Price: 3489 <br /> Clarity: VS2","Carat: 1.03 <br /> Price: 5087 <br /> Clarity: SI1","Carat: 1.57 <br /> Price: 12320 <br /> Clarity: SI1","Carat: 0.37 <br /> Price: 817 <br /> Clarity: VS1","Carat: 2.17 <br /> Price: 17747 <br /> Clarity: SI1","Carat: 0.67 <br /> Price: 2427 <br /> Clarity: VS1","Carat: 0.27 <br /> Price: 603 <br /> Clarity: VS1","Carat: 1.21 <br /> Price: 9541 <br /> Clarity: VVS2","Carat: 1.52 <br /> Price: 13034 <br /> Clarity: VS1","Carat: 0.3 <br /> Price: 886 <br /> Clarity: IF","Carat: 0.31 <br /> Price: 1046 <br /> Clarity: VVS1","Carat: 1.5 <br /> Price: 10428 <br /> Clarity: SI1","Carat: 0.32 <br /> Price: 688 <br /> Clarity: VS2","Carat: 0.31 <br /> Price: 462 <br /> Clarity: SI2","Carat: 0.4 <br /> Price: 912 <br /> Clarity: VS2","Carat: 1.47 <br /> Price: 8314 <br /> Clarity: SI2","Carat: 1.51 <br /> Price: 7455 <br /> Clarity: VS2"],"type":"scatter","marker":{"colorbar":{"title":"carat","ticklen":2},"cmin":0.22,"cmax":3.02,"colorscale":[["0","rgba(68,1,84,1)"],["0.0416666666666667","rgba(70,19,97,1)"],["0.0833333333333333","rgba(72,32,111,1)"],["0.125","rgba(71,45,122,1)"],["0.166666666666667","rgba(68,58,128,1)"],["0.208333333333333","rgba(64,70,135,1)"],["0.25","rgba(60,82,138,1)"],["0.291666666666667","rgba(56,93,140,1)"],["0.333333333333333","rgba(49,104,142,1)"],["0.375","rgba(46,114,142,1)"],["0.416666666666667","rgba(42,123,142,1)"],["0.458333333333333","rgba(38,133,141,1)"],["0.5","rgba(37,144,140,1)"],["0.541666666666667","rgba(33,154,138,1)"],["0.583333333333333","rgba(39,164,133,1)"],["0.625","rgba(47,174,127,1)"],["0.666666666666667","rgba(53,183,121,1)"],["0.708333333333333","rgba(79,191,110,1)"],["0.75","rgba(98,199,98,1)"],["0.791666666666667","rgba(119,207,85,1)"],["0.833333333333333","rgba(147,214,70,1)"],["0.875","rgba(172,220,52,1)"],["0.916666666666667","rgba(199,225,42,1)"],["0.958333333333333","rgba(226,228,40,1)"],["1","rgba(253,231,37,1)"]],"showscale":false,"color":[0.73,0.7,0.31,0.31,0.31,0.83,0.51,0.7,0.4,1.1,0.52,0.32,1,1.01,2.01,1.6,1.01,0.31,0.38,1.03,1.31,0.7,2.01,0.8,1.2,0.31,0.82,0.41,0.33,0.33,1.3,0.35,1.25,0.62,0.3,0.4,0.9,0.4,0.5,1.09,0.91,0.34,1.66,0.42,1.17,1.27,0.9,1.55,1.5,1.06,0.9,0.42,0.32,0.38,0.78,0.3,2.01,0.34,1.31,0.9,0.33,0.31,1.22,2.15,1.27,1.06,2,2.01,0.23,1.16,1.01,0.34,0.55,0.43,0.5,0.3,1.52,1.22,0.3,0.5,1.26,0.41,0.31,0.5,0.39,0.31,1.3,1.05,1.06,0.51,0.52,1.52,0.31,1.33,0.41,0.4,0.88,0.32,0.7,0.53,0.52,0.69,0.65,1.2,1,1.2,1.5,0.38,1.51,0.35,0.3,1.01,0.54,0.31,0.5,1.04,1.19,2.13,1.1,0.3,1,0.71,1.02,0.51,0.71,0.55,0.34,0.79,0.36,0.3,0.89,0.57,1.22,1.09,0.54,1.02,0.9,1.14,0.26,0.54,0.9,1,2.02,1.5,0.53,1,2.3,1,1.51,0.62,0.81,0.33,2,0.31,1.01,0.57,0.77,1.03,1.23,1.24,0.71,0.31,1.04,1.19,0.41,0.4,1,0.53,0.27,1.11,1.63,0.9,0.42,1.73,0.41,1.3,0.24,0.35,0.3,1.01,0.27,1.16,0.59,0.7,0.5,1.5,0.55,0.36,1.05,0.3,0.9,1,0.51,0.81,0.72,0.33,0.23,0.51,0.84,0.59,0.91,0.52,0.87,0.3,0.96,0.63,1.09,0.92,1.01,0.3,0.31,1,2.2,1.01,0.71,0.72,1.31,0.77,0.51,0.59,1.54,0.57,0.71,0.51,1.31,1.01,1,1.5,1.06,0.32,0.34,0.31,1.11,0.41,0.72,0.32,0.53,0.74,0.4,0.7,1.5,1.13,0.5,0.41,1.04,0.92,1.5,0.9,0.31,1.14,0.58,1.57,0.91,0.7,0.7,0.92,0.3,2,1.32,0.58,1.05,0.7,0.35,0.7,1.5,0.58,1.01,0.31,0.32,0.7,1.52,0.7,0.7,0.92,0.9,0.34,0.33,1.5,0.42,1.05,0.31,0.7,0.32,1,1.02,0.27,1.08,0.43,1.13,1.5,0.3,0.56,1.2,0.36,0.33,0.51,2.07,1.01,0.33,0.6,0.6,0.35,0.75,1.12,0.26,1.01,0.43,0.27,1,1.03,1.01,0.33,0.24,1.01,0.3,1.02,1.5,0.5,0.3,0.71,1.11,0.28,1.29,0.57,0.33,0.75,0.35,0.32,0.32,0.29,1.03,1.7,0.9,0.32,0.61,0.51,1.13,1.02,0.3,0.31,1.51,0.81,0.7,0.74,0.51,0.35,0.62,0.32,0.58,1.01,1.07,0.7,1.04,0.51,0.53,0.3,0.9,0.78,0.33,1.51,1.02,0.43,1,0.38,0.6,0.54,0.49,0.9,1.23,1.21,0.7,0.31,0.56,1.71,0.25,0.35,0.32,0.4,0.82,0.9,0.41,0.3,1.5,0.76,0.51,0.4,0.7,0.24,0.31,0.31,0.73,0.41,0.28,1.17,1.5,1.5,1.26,1.18,1.02,0.26,0.4,1,0.35,1.01,0.34,0.73,1,0.45,0.71,2.12,0.3,0.4,0.32,0.31,0.33,1.01,1.52,1.77,1.08,0.33,0.32,0.27,0.32,1.24,0.36,0.71,0.9,1.47,0.56,1.8,2.02,0.3,0.38,0.31,0.91,0.39,0.49,0.31,0.55,0.33,0.91,0.73,0.42,1.3,1.24,0.72,1.01,1.72,0.72,1.21,0.33,0.71,0.7,0.73,1.5,1.01,2,1.5,0.71,0.34,1.29,1,0.72,0.3,1.17,0.5,0.7,0.3,0.31,2.04,1.33,0.32,1,1.52,0.71,0.3,0.33,0.34,0.34,1.01,1.52,2.02,1.64,0.4,1,0.38,0.3,1.1,1.08,1.02,0.84,1.51,2.01,0.71,0.32,0.32,1.19,1.01,0.71,0.31,0.41,0.7,1.25,0.87,1.5,0.59,1.01,0.36,0.31,0.78,0.7,0.39,0.7,1.07,0.4,1.04,0.36,0.81,0.52,0.31,1.1,0.41,1.07,0.73,0.23,0.32,0.26,0.3,0.33,0.4,1,0.73,0.71,0.38,0.39,0.41,0.35,0.55,1.01,0.42,0.31,0.43,1.12,0.51,0.52,0.3,0.44,1.01,0.9,1.56,1.01,0.33,0.3,0.9,1.7,2.02,2.17,0.28,0.53,1.59,0.5,1.07,0.53,0.42,0.33,1.08,0.32,0.44,0.73,0.84,0.41,0.32,0.31,0.3,1.51,0.5,1.04,0.34,0.31,0.41,1.51,0.91,1.22,1.2,0.51,1.51,0.81,0.31,1.24,1.01,0.36,0.7,1.04,0.5,1.02,0.31,0.41,1.12,0.42,0.27,0.68,2.14,0.53,0.7,0.31,0.7,0.34,0.41,0.57,1,0.32,0.74,0.9,0.53,1.3,1,0.5,0.95,2.01,0.51,1.11,0.75,1.5,0.72,0.74,0.7,1.01,0.24,1.01,0.43,0.43,0.41,1.24,2.03,2.01,1.23,0.78,0.4,0.63,2.32,1.12,0.56,1.59,1.04,1.01,0.32,0.7,0.42,1.5,0.32,1,2.04,1.05,1.53,1.48,1.12,1.01,0.82,1.51,0.56,0.3,0.25,0.93,1.08,0.6,1.53,1.5,0.4,0.9,1.03,1.23,0.84,0.71,0.26,0.37,0.52,2.11,1.51,1,0.55,0.51,1.01,0.32,0.58,1.26,0.78,0.84,0.23,1.5,0.33,0.51,0.73,0.52,2.04,1.54,1.53,0.24,0.55,0.52,0.44,1.19,0.35,1.76,1.01,0.41,0.76,0.5,1.09,1.21,0.52,0.39,0.5,0.92,0.37,0.7,1.71,1.21,0.52,0.51,1.2,0.31,0.41,0.81,0.72,0.83,0.63,1.8,0.32,0.34,2.14,0.7,0.3,1.21,0.54,0.41,0.43,0.32,0.24,1,0.51,0.8,1.54,0.74,1.23,0.32,0.29,1,0.41,0.41,0.55,0.57,0.39,0.4,0.82,1.5,0.7,0.34,1.03,1.03,1.04,0.61,0.31,0.72,0.98,0.29,0.27,1,0.38,1.83,0.51,1.16,0.5,1.52,0.91,1.04,1.02,1.01,0.7,0.31,0.4,0.65,0.7,0.3,1.02,0.32,0.71,0.33,0.38,0.51,0.3,0.54,1,0.3,0.31,1.51,1.02,0.31,0.76,1.12,0.71,0.55,1.12,0.58,0.41,1.02,0.71,1.05,0.33,0.7,1,0.3,1,1.5,0.86,0.26,0.35,0.5,1.5,1.01,0.32,1,0.39,0.41,1.2,1,1.01,0.73,0.31,1,0.33,0.58,0.93,1.31,1.04,0.42,1.04,1.02,0.79,0.38,1.01,1.01,0.33,0.67,0.44,1.23,0.52,0.7,0.8,0.33,1,0.37,0.74,1.2,0.31,0.71,0.51,0.76,1.27,1.7,0.31,1.53,0.83,0.52,1.06,0.53,1.51,1.51,0.71,0.34,1.52,0.9,1.54,0.56,1,1.01,0.32,1.2,0.75,0.62,1.16,0.82,0.3,1.52,0.91,0.37,0.32,1.11,1,1.01,1.5,0.58,1.06,1.5,0.32,0.91,0.51,0.31,0.38,0.27,0.9,0.78,0.55,0.44,0.7,0.54,1.04,1.04,0.7,0.43,1.04,0.32,0.8,0.38,0.5,1.51,0.5,1.19,0.94,0.45,0.32,0.4,0.25,0.79,1.52,1.22,0.43,0.31,1.08,0.43,0.7,1.21,0.3,1.22,1.21,1,0.74,1.5,1.64,0.93,0.31,0.91,0.37,1.21,0.73,0.3,2.02,1.21,0.4,1.06,0.3,1.57,0.72,0.39,0.41,0.38,0.31,0.65,1.32,0.9,0.32,1.2,0.46,0.77,1.01,1.12,1.56,0.3,0.72,1.02,1,2.22,0.53,1.22,0.56,0.34,2.24,0.33,1.05,0.31,0.73,2.09,1.09,0.7,0.57,0.73,0.33,0.3,0.33,1.73,0.51,0.31,0.57,0.8,0.71,0.32,1.25,0.34,0.32,0.68,1.51,0.52,0.3,0.52,0.32,1.23,1,0.71,0.33,1.58,0.7,1.27,0.37,0.56,1.5,1.52,0.97,0.77,0.72,0.31,0.41,1.02,1.03,1.74,1.01,0.3,0.41,0.7,1.24,1,1,0.6,1.31,0.32,0.7,0.38,1.59,1.01,0.38,0.55,2.02,0.24,0.34,0.9,0.31,0.31,1,0.97,0.4,0.62,1.03,1,0.28,0.48,1.01,1,0.43,1.01,0.38,0.3,0.7,0.91,0.66,1.01,0.32,0.33,1.56,0.79,1.01,0.71,2.01,0.5,1.04,1.21,0.41,1.16,0.32,0.58,0.38,0.34,0.5,0.32,1.15,1.05,0.54,0.3,1.29,0.4,0.33,1.03,0.7,1.2,0.3,0.33,0.5,0.6,0.62,1.07,1.13,0.71,0.32,0.91,1.13,0.9,0.71,0.3,1.69,0.58,1.59,0.28,0.53,0.39,2.16,0.31,1.55,1.6,0.4,1.01,1.03,0.92,0.62,0.3,0.4,0.7,0.9,0.82,0.5,0.92,0.31,0.91,0.4,0.4,0.26,1.11,2.48,0.32,1.09,0.42,2.01,2.02,0.7,1.01,0.32,0.3,0.7,0.38,0.31,2.09,0.41,0.3,0.9,0.7,0.28,0.9,0.54,0.28,0.31,0.93,2.02,0.41,1.37,0.37,2.11,0.23,0.51,1.09,1.18,1.17,0.67,1.6,0.76,1.04,2.01,1.06,1.24,1.06,1.16,1.13,0.51,0.5,1,1.08,1.55,0.41,0.91,0.4,1.07,0.57,1.57,0.71,0.31,0.3,0.35,0.45,0.52,0.38,1.23,0.32,0.51,0.39,0.9,1.18,1.53,0.3,0.76,0.51,0.42,0.41,1.05,0.52,1,0.28,0.23,1.01,0.77,1.51,1.7,1,0.32,1.11,0.85,1,1.08,1.5,0.51,1.5,1.26,1.01,0.31,0.53,0.41,0.53,0.43,0.51,0.8,0.71,0.34,0.9,0.5,1.51,2.01,1.64,0.33,1.04,0.78,0.76,0.52,0.44,1.11,0.9,0.56,0.79,0.71,0.73,0.4,1.03,0.32,0.52,1,1.33,1.54,1.7,1.01,0.57,1.21,0.32,1.01,1.04,0.5,0.32,1,1.3,0.31,0.5,0.43,1.24,0.32,0.53,1.15,1.21,1.04,0.27,1.04,0.71,0.27,0.32,1.5,1.01,1.02,0.56,2.06,1.2,0.73,0.32,0.73,0.9,0.25,1.01,1.09,0.41,0.31,0.53,0.52,0.58,0.38,0.73,0.35,2.04,0.32,0.75,0.5,0.72,0.5,0.32,0.53,0.72,0.79,0.35,0.7,0.59,0.33,1.45,0.3,0.32,0.31,0.45,0.32,0.31,0.7,0.96,1.5,1.01,0.34,1.2,0.43,1.04,1.13,1.22,1.09,1.29,0.4,1,0.39,0.41,0.9,1.01,0.72,1.02,1.02,0.79,2.16,0.37,0.3,0.42,0.41,0.56,1.29,0.31,0.4,0.35,1.22,0.4,0.5,1.01,0.32,0.92,0.5,0.31,0.34,0.7,2.08,0.9,0.34,0.42,0.51,0.45,2.12,0.26,0.32,0.33,0.77,0.32,0.9,1.2,1.03,0.26,0.53,1.01,0.3,0.42,0.3,1,1.01,1.51,0.33,1.31,0.39,0.51,0.9,1.01,0.39,0.32,1.12,0.53,0.5,1.5,0.32,1.45,0.78,0.3,0.4,1,2.1,1.5,1.12,1.26,1.6,0.4,0.31,0.55,1.36,0.81,1.01,2.05,0.7,0.38,1.82,0.5,0.32,1.44,0.52,1,1.07,0.8,0.72,2.03,1.38,0.4,0.74,1,2.01,1.01,1.54,0.26,0.44,0.3,0.31,0.5,0.59,2.05,1.02,0.98,0.4,0.7,0.5,0.37,0.7,0.32,0.4,0.55,0.57,0.46,0.55,0.71,1.14,0.41,1,0.3,1.1,0.5,2.01,0.4,0.34,0.3,0.34,1.51,0.45,1.08,1.13,0.3,0.7,0.34,1.14,1.07,0.43,1.2,0.5,0.41,0.31,0.57,0.93,2.03,1.01,1.07,0.73,0.52,0.53,0.37,0.71,1.12,1.51,0.32,1.87,0.41,0.7,1.09,0.38,1.24,1.2,1.14,0.62,0.7,1.01,1.01,1.1,0.5,1.53,0.31,1.01,0.55,1.5,0.32,1.22,0.31,1.11,1.13,0.53,1.03,1.06,1.04,0.72,1.05,0.3,0.7,1.59,0.34,0.73,0.41,0.81,0.73,1.21,1,1.07,0.52,0.74,0.9,1,1.03,0.41,0.39,1.27,0.33,0.34,0.33,1.22,0.52,0.71,0.31,0.8,2.06,0.51,0.31,0.5,0.32,1.01,1.02,0.34,0.77,0.32,1.21,1.47,0.72,2.42,1.01,0.72,0.33,0.7,0.7,0.4,0.51,0.5,1.59,1.2,0.61,1.01,0.7,1.13,2.02,1.23,0.31,0.71,0.3,1.17,0.32,0.92,0.32,2.19,0.53,0.3,0.77,0.32,0.32,0.3,0.96,0.42,0.35,0.31,0.81,0.53,1.2,2.01,0.41,1.54,0.95,1.61,1.04,0.34,0.3,0.51,0.8,1.17,1,0.51,0.32,0.41,1.1,2.39,1.7,1.28,1,0.52,2.04,1.01,1.01,1.5,0.53,2.01,0.71,0.31,0.34,0.42,0.36,0.3,0.41,0.6,0.33,0.35,0.36,1,1.05,0.41,0.32,0.56,1.18,0.49,1.05,1.11,0.52,1.14,0.36,0.52,1.35,0.5,0.91,0.52,0.62,1.51,0.71,0.4,0.48,0.91,1.57,0.55,1.51,0.4,1.24,0.3,0.32,0.3,0.7,0.34,0.22,1.56,0.53,1.01,0.56,1.25,1.01,0.79,0.43,0.5,1.01,1.59,1.2,0.28,1.09,0.76,0.9,0.4,0.54,1.51,0.77,0.79,0.5,0.23,0.78,0.63,0.91,0.36,0.33,0.31,0.38,0.38,1.21,0.33,2.03,1.01,0.33,2.18,1.54,0.41,0.33,0.7,1.12,0.51,1.16,0.91,1.5,0.3,2.01,0.33,0.55,0.3,0.4,0.71,1.5,0.91,0.5,0.7,0.52,0.39,0.28,1.58,1.34,0.91,0.41,0.33,0.56,1.05,1.03,0.53,0.93,0.31,1,1.26,0.42,1.06,0.38,1.41,0.4,1.51,1.52,1.08,1.5,0.44,0.3,1.27,0.3,1.1,1.54,0.9,0.4,0.35,1.09,0.71,0.57,1.5,2.09,0.3,2.01,0.5,0.41,0.82,0.42,0.7,0.31,0.59,0.55,1.5,1.03,1.01,0.35,0.56,0.7,0.9,0.38,0.72,1.13,0.61,0.32,0.78,1.03,0.23,1.01,0.71,0.37,1.21,0.5,0.53,2.21,0.7,0.45,0.66,0.33,0.52,0.3,2.04,0.36,0.34,1.01,1.5,0.24,0.34,0.71,1.07,1,0.27,2.14,2.24,0.7,1.19,1.5,0.56,0.79,0.77,0.34,1.01,0.3,0.71,1.01,0.34,0.7,0.3,1.03,0.57,1.13,0.71,0.62,0.41,0.3,0.71,0.92,1.24,0.53,1.01,0.4,0.7,1.02,0.59,0.72,2.03,0.57,0.35,1.06,1.21,0.76,0.42,0.4,1.1,0.77,1.2,1.34,0.6,0.77,1.52,1.23,0.72,0.3,0.42,1.52,2.11,0.58,0.36,0.51,0.93,0.64,0.94,1.7,0.25,0.32,0.56,0.95,0.29,0.33,0.41,1,1.02,0.3,2.05,0.7,0.55,1.58,1.01,2.03,0.5,0.32,0.38,1.08,0.38,0.59,0.9,1.03,0.78,1.5,1.54,2.05,0.53,1.24,0.45,0.51,0.25,0.36,0.58,0.51,0.9,0.7,0.3,0.4,0.41,0.31,0.53,0.25,0.55,0.41,0.7,0.73,0.32,0.5,0.33,0.31,1.51,0.8,1.41,0.34,0.41,0.7,0.39,0.29,2.38,0.9,0.36,0.7,0.25,1.01,1.16,1.01,0.7,0.3,0.9,0.4,1.6,0.3,0.9,0.7,0.51,0.3,1,0.55,0.5,0.31,0.57,1.01,1.02,0.5,0.25,0.72,1.23,1.26,0.52,0.7,0.32,1.28,0.7,0.53,0.3,1.88,0.9,1.52,1.4,0.32,1.04,0.9,2.1,2,0.36,0.33,0.32,0.54,0.4,1.09,1.01,1.03,0.78,0.42,0.34,1.06,0.32,0.71,0.31,0.37,0.34,0.7,0.35,0.43,0.5,1,0.9,0.28,0.41,1.18,1.02,1.2,0.74,1.2,0.7,0.53,0.26,0.32,1.15,1.07,0.52,1.04,1.01,0.7,0.56,1.01,0.32,1.11,1,0.51,1.7,1.43,0.55,0.51,1.51,0.74,0.9,0.37,1.01,1.04,0.31,1.01,2.07,0.31,0.31,0.8,0.72,0.82,0.31,0.39,2.08,0.42,0.34,0.36,0.6,0.9,0.74,0.74,1.48,0.33,0.37,0.71,0.95,0.73,0.37,1.5,1.13,0.4,1,0.85,0.51,1.01,1.02,1.54,0.92,0.76,1.04,0.9,0.37,1.02,1,0.3,0.36,0.41,0.71,0.5,0.34,1.74,0.3,0.4,1.2,1.21,0.57,1.12,0.58,0.7,1.07,0.7,0.7,1.7,1.01,0.33,0.52,0.27,0.4,0.42,0.25,0.37,1.14,0.3,0.34,0.36,0.31,0.91,1.17,0.34,0.33,0.3,0.42,0.3,1.01,0.4,0.41,0.3,0.32,1.02,2.01,0.3,0.31,0.7,1.02,1.04,1.1,1.24,0.61,2.14,1.02,0.31,1.15,0.4,0.26,0.34,0.8,1.21,0.33,0.31,1.51,1.34,0.53,1.17,0.3,2,0.3,2.1,0.32,0.5,0.53,0.31,0.8,0.32,2.01,1.11,0.36,0.72,1.25,1.2,0.73,1.07,0.51,0.58,0.73,1.5,0.53,0.7,0.43,1.5,0.52,1.25,2.1,0.31,0.6,1.01,0.31,0.45,0.83,0.5,0.41,2.5,2,0.7,1.13,0.31,1.01,0.83,0.41,0.5,1.51,1.3,0.71,0.3,0.71,0.46,1.03,0.37,2.51,1.07,1,0.7,0.25,0.34,0.3,0.55,0.4,0.51,0.3,0.51,0.42,0.51,1.11,0.4,0.43,0.36,1.16,0.52,1.03,0.33,0.41,0.53,0.27,0.38,0.34,0.41,0.3,0.3,0.46,1.56,0.41,0.8,0.24,1.86,0.57,0.28,1.04,0.32,0.31,1.1,0.9,0.57,0.3,0.66,0.32,0.52,1.34,0.3,1.22,0.3,2.51,1.52,1.01,1,0.34,0.52,0.46,0.52,0.96,0.4,1.62,0.32,1.07,1.74,0.35,1.74,1.01,1.01,0.71,0.75,0.32,0.74,0.71,1.01,2.08,0.34,0.75,1.01,0.31,1.24,0.31,0.69,1.24,0.53,0.24,1.01,1.3,1.09,0.75,0.32,0.7,1.21,0.3,0.71,0.38,0.5,2.02,1.19,0.25,0.33,0.41,0.34,0.34,0.3,0.41,0.93,0.45,0.71,0.38,1.57,0.32,1.55,0.9,1.2,0.76,0.6,0.95,0.73,2.3,0.3,0.3,1.54,0.51,0.55,0.31,0.26,0.9,0.5,1.01,0.93,0.5,0.41,2.01,1.1,0.54,0.41,1.2,0.34,1.7,0.77,1.11,0.9,1.02,2.6,0.83,1.15,0.3,0.34,1.65,1.51,1.26,0.37,0.72,0.47,0.74,0.7,0.31,0.71,0.4,0.3,0.54,0.3,0.32,0.36,0.54,0.32,1.09,1.18,0.35,1.29,0.3,0.41,0.3,0.7,0.37,0.24,0.41,2.04,0.32,0.71,0.3,0.71,0.51,0.56,0.27,0.27,1.01,2.01,0.3,0.54,0.39,0.38,0.91,1.01,0.92,0.62,2.44,1,0.38,0.57,1,1,0.51,0.31,0.94,0.76,1,0.93,0.84,0.34,0.72,0.31,0.23,1.16,0.5,0.5,1.66,1.01,1.58,0.57,1.56,2,0.7,0.4,1.02,0.73,0.32,0.55,1.03,0.41,0.33,1.35,1.51,0.53,1.06,1.2,1.25,1.01,0.51,0.7,0.71,1.03,0.31,0.36,0.36,0.32,0.3,1.29,0.31,1,1.04,2.02,0.42,0.53,0.3,0.53,0.55,0.3,0.3,0.29,0.62,1.05,1.51,0.32,0.62,0.37,0.32,0.3,0.42,0.57,1.51,0.98,0.56,1.27,0.3,1,0.31,0.32,1.27,0.55,0.31,1.16,0.39,0.41,0.42,2,0.55,1.59,0.31,0.9,0.43,0.31,0.9,0.42,0.85,0.23,0.8,0.32,1.01,0.59,0.7,0.27,0.64,1.08,0.26,0.43,0.33,1.23,1.51,0.94,1.23,0.3,0.31,2.19,0.38,0.9,0.34,0.35,0.33,1.51,0.32,0.5,1.01,0.91,1.51,1,1.13,0.5,0.8,0.36,0.36,1.25,1.58,0.32,0.3,0.8,0.39,0.42,0.42,0.7,0.54,0.42,0.74,1.02,0.31,0.36,2.02,0.91,1.57,0.33,1.05,1.01,0.51,1.02,0.7,1.18,1.12,0.32,2.13,0.3,1.02,0.38,0.72,2.01,0.41,1.25,1.04,1.18,1,0.72,1.2,0.55,1,1.05,1.51,0.31,0.75,0.56,1.01,0.41,0.7,1.01,0.71,1.2,1,1.51,0.9,1.01,0.43,0.46,1.5,0.41,0.42,1.51,0.5,1.02,0.75,0.53,0.8,0.7,1.01,0.9,0.61,0.55,0.31,0.52,0.31,0.74,0.41,0.31,0.56,0.36,0.5,1.8,0.71,0.52,1.01,0.53,2,2.16,0.53,0.32,0.36,0.84,0.7,0.31,0.7,0.4,1.02,1,1.33,1,1.08,0.53,0.3,0.5,1.01,0.8,0.54,0.81,0.56,1.01,0.32,0.54,0.3,0.3,0.4,1.01,1.17,0.35,0.32,1.51,0.4,1.6,0.31,0.31,0.99,2.01,0.27,0.3,0.7,0.67,0.24,0.71,0.54,0.71,2.09,0.32,1.01,1.06,0.7,0.3,2.1,0.51,0.54,1.03,1.51,0.34,0.41,0.46,1.04,0.33,0.51,0.34,0.31,1.23,0.33,0.34,0.31,0.65,1.58,0.23,2.02,1.83,0.91,0.31,1.02,1,0.36,1.51,1.34,0.92,1.16,0.3,1.01,2.15,0.9,0.9,1.33,0.54,0.33,0.31,1.2,0.41,0.51,1.85,0.32,1.32,0.76,0.23,0.54,0.32,1.02,0.5,1.02,0.44,1.01,1.23,0.9,1.53,1.3,0.73,0.31,0.34,0.27,1.63,0.5,0.74,1.16,0.3,2,0.3,1.32,0.56,0.38,0.32,1.12,1.2,0.53,0.37,1.61,0.79,1.51,1.04,1.01,1.53,0.3,0.3,1.03,0.3,1.05,0.31,0.3,0.31,0.36,0.51,0.75,0.71,0.38,0.43,1.06,0.72,1.25,0.9,0.53,1.21,0.83,1.52,0.3,1.5,1.21,0.77,0.39,0.4,0.32,0.92,0.36,1.02,0.25,0.3,0.3,1.5,1.01,1.01,1.06,0.9,0.7,0.5,1.59,1.18,1.11,1.04,0.74,0.28,1.08,0.52,0.36,1.03,0.36,2,0.7,0.77,0.7,0.31,0.35,0.42,0.67,0.56,2,0.9,0.42,0.33,0.6,0.41,1.01,0.83,0.3,0.34,0.35,0.76,0.9,2,1.51,0.7,0.3,0.7,1.01,1.13,1.52,0.4,0.31,1.09,0.52,0.39,1.39,0.41,0.7,1.01,1.1,0.32,0.35,0.31,0.3,1.52,0.9,0.37,0.71,1.64,0.59,1.09,2.01,0.41,1.26,0.76,0.35,0.7,1.51,0.28,0.9,0.96,1.03,1.51,1.1,1.09,1.03,1.06,0.31,1.53,0.52,0.7,1.5,1,1.07,0.31,0.33,0.4,0.32,0.91,0.5,0.32,0.37,1.1,0.34,0.51,0.4,1.14,1.05,0.32,1,0.51,1.02,0.32,0.32,0.3,0.71,1.01,0.39,1.52,0.44,0.54,0.72,1.01,1.04,0.9,0.3,1.7,1.01,0.93,0.5,0.25,0.3,0.71,0.41,1.31,1.16,0.71,0.3,1,1.13,0.34,0.68,0.4,0.53,0.7,0.41,1.61,1.21,0.32,0.63,1.13,0.71,0.23,1.01,0.81,1,1.22,1.53,0.32,0.33,0.39,1.08,0.92,0.72,0.54,0.73,0.36,0.71,0.7,0.34,0.41,0.72,0.31,1,1.77,0.9,2.01,0.54,1.21,1.7,0.44,0.9,1.22,1.55,0.81,0.5,1.3,0.7,1.02,0.5,0.33,0.41,0.76,0.61,1.2,0.58,0.27,1.2,1.01,0.51,0.31,0.4,0.71,1.56,0.41,0.4,2.36,1.55,0.24,0.51,1.1,1.02,1,0.5,1.5,0.73,0.31,0.56,0.38,0.54,0.38,0.77,0.51,0.73,0.71,0.42,1.51,0.52,1.55,0.76,1.01,0.73,0.91,0.39,0.33,0.37,1.01,0.41,1.12,1.15,0.42,0.42,0.3,0.38,1.2,0.42,0.7,0.71,2,0.48,0.29,0.38,0.9,1.3,0.33,2.52,0.33,0.62,0.31,0.54,1.04,0.4,0.42,0.51,0.92,0.5,0.41,0.7,0.7,1.59,0.3,1.51,1.04,0.31,1.05,1,2.01,0.55,0.71,0.23,1.5,0.71,0.71,2,0.7,0.79,2.28,0.37,1.27,0.36,1,0.82,0.42,0.57,1.26,1.39,1,1.01,1.15,0.33,0.24,0.7,1.34,0.37,0.31,2.2,1.08,1,0.52,1.01,1,0.75,0.41,0.24,1.5,1.51,0.8,1,0.54,1.07,1.08,1.5,0.41,1.5,0.7,0.37,0.8,1.02,0.5,0.78,0.31,1.06,0.5,1.15,0.25,2.26,1.05,0.57,1.01,0.32,0.53,0.91,0.41,1.23,1.31,0.9,0.42,1.13,0.34,1.05,0.58,0.3,1,0.42,0.91,0.32,0.86,1.33,1.52,0.72,0.7,0.53,0.5,0.42,1.03,1.16,2.04,0.4,1.01,0.5,1.04,0.75,1.26,1.06,0.34,0.32,1.01,0.91,0.36,0.9,1.2,0.9,1.23,0.75,0.5,0.53,0.3,0.91,0.51,2,0.75,0.9,1.42,0.34,1.3,1.51,1.01,1.27,2.01,0.4,0.27,1.03,0.73,2.02,0.38,0.36,1.51,1,1.51,0.32,1.09,1.34,0.4,1.04,0.7,0.3,0.31,0.76,0.27,0.33,0.64,0.39,0.7,1.02,1,0.72,0.7,1.23,0.74,0.3,0.42,0.71,0.76,0.38,0.9,1.02,1.67,0.51,0.9,1.2,0.9,1.53,1.11,0.41,1.32,0.38,0.35,1.52,0.9,0.24,0.75,0.91,0.4,0.33,1.31,0.36,2.01,0.33,1.51,1.25,1.02,1.22,0.34,0.33,0.7,0.77,0.5,0.31,0.3,0.56,2.02,1.02,1.01,1.51,2.04,0.71,1.57,0.35,0.53,0.32,0.78,0.53,2.1,0.82,0.71,0.3,1.05,0.7,0.33,2.01,2.01,1.01,0.42,1.02,0.93,0.81,2.61,1.18,0.7,0.28,0.39,0.71,1.59,1.04,0.53,0.9,0.32,1.18,0.31,0.31,0.3,0.4,1.54,1.11,0.57,1.5,1.65,0.72,0.29,0.52,0.76,1.06,1.62,0.71,1.21,0.52,1.12,0.96,1.01,0.3,0.32,1.51,0.3,1.16,0.66,0.79,0.59,0.57,1.01,1.26,0.44,0.5,1.5,0.51,1.2,0.71,0.9,1.06,1.24,0.53,0.72,1.01,0.36,1.14,0.3,1.58,1.11,1.21,0.3,0.33,0.81,0.39,0.51,1.01,0.42,0.39,0.92,1,1.57,0.32,0.91,0.44,0.53,0.58,0.32,0.7,0.38,0.3,0.7,0.77,2.01,0.71,1.71,0.42,2.06,1.98,1.01,0.54,0.5,1.04,0.36,0.61,0.24,0.91,1,0.31,0.31,1.73,0.3,0.31,0.42,1.01,0.52,1,1.27,1.22,0.54,0.31,0.98,0.84,0.31,0.33,0.3,2.5,1.52,0.38,0.75,0.43,1.02,0.77,0.35,0.33,0.5,1.02,0.72,0.41,0.38,0.7,0.41,1.01,0.31,1.08,2.02,1.56,0.73,1.01,0.8,0.35,0.71,0.41,0.86,0.9,1.46,0.4,1.22,0.53,0.71,1.52,0.3,0.42,0.42,1.41,0.3,1.51,0.5,1.68,1.53,0.54,1.14,2.4,0.42,1.03,0.9,1.05,1,0.41,1,0.7,0.3,0.54,1.01,0.71,0.7,1.01,0.4,0.58,0.38,1.08,0.32,0.34,0.3,0.32,1.51,0.33,0.71,0.48,1.09,0.72,1.67,0.75,0.27,0.41,0.55,0.46,0.34,0.7,0.54,1.01,2,0.4,1.07,0.51,0.51,1.51,1.01,1.02,0.31,0.3,0.3,1,0.36,0.51,1.02,0.7,0.58,0.33,1.01,1,0.76,0.3,2.01,0.9,0.35,0.52,0.34,1.03,1.03,0.29,0.36,0.42,0.3,0.55,0.5,1,0.31,0.72,0.54,0.3,1.56,0.9,1.11,0.83,0.28,0.43,0.55,0.33,1.02,1.01,0.57,1.01,0.55,0.33,0.9,0.33,0.54,0.41,1.15,1.03,0.41,0.59,0.31,1.62,0.3,1.37,1.01,0.31,0.32,0.58,1.54,1.13,0.8,0.4,0.96,0.56,1,0.5,0.3,0.51,1.5,1.01,0.58,0.53,0.38,0.56,0.28,0.53,0.86,0.59,0.27,0.73,0.6,0.35,0.57,1.5,1.13,1.51,0.81,0.23,0.3,0.78,0.25,1.5,0.93,0.52,1.23,0.73,0.32,0.72,1.5,1.12,1.5,0.42,0.3,0.34,0.92,0.43,1.1,0.87,0.96,0.71,1.23,0.24,0.6,0.7,0.3,1.2,0.34,1.6,1.52,1.09,1.2,1.59,0.24,0.41,0.54,1.31,0.32,0.35,0.31,1,1.95,0.32,0.4,0.71,0.7,1.15,1.21,0.52,0.7,0.82,0.26,0.92,1.11,0.43,2.2,1.38,0.51,1.01,1.01,2.03,0.52,1.21,0.44,1,1.04,1.29,0.41,1.08,0.71,1.65,0.52,1.11,0.32,0.74,0.3,0.3,0.41,0.63,1,0.7,1.16,0.4,0.69,1.01,0.33,0.5,1.79,1.05,1.09,0.79,0.51,0.35,1.02,1.06,1.01,0.5,1.51,0.3,0.31,1.2,1.03,0.41,1.11,0.33,0.54,0.32,1.26,0.46,0.44,0.33,0.58,0.3,0.5,1.15,0.76,1.5,0.51,2,0.72,1.12,0.7,0.7,0.32,1.04,0.81,0.44,0.38,0.32,2.03,0.39,1.34,0.77,0.51,0.33,0.9,1.53,1,0.38,1,1,1.7,0.55,1.13,0.57,0.31,0.41,0.9,0.32,0.55,0.91,1.5,0.33,0.35,1.5,1.01,0.3,0.34,0.33,1.01,1.04,0.41,0.71,1.05,1.07,2.02,0.34,0.73,0.71,1.24,0.26,0.31,0.38,0.34,1.2,0.5,0.42,1.75,0.61,0.36,0.53,0.9,1.36,0.24,1.23,0.31,1.01,0.36,0.3,1.05,0.32,2.02,1.21,0.65,0.7,0.41,0.43,0.41,0.4,0.55,0.52,1.02,1.02,0.35,0.31,1.03,0.72,0.41,0.71,0.47,0.31,2.01,0.5,0.34,1.23,1.14,1.53,1.03,1.71,1.33,1.04,0.3,0.33,1,0.3,1.02,0.54,0.83,1.05,0.31,0.32,2.04,0.73,1.01,1,0.3,0.44,0.53,1.71,1.02,0.61,0.71,1.41,1.11,0.52,0.74,0.9,0.31,0.32,1,0.7,1,0.72,0.4,1.59,0.83,1.61,0.3,1.26,0.56,0.38,1.12,0.74,0.31,0.42,0.83,0.5,0.81,0.71,0.31,0.7,0.53,0.33,0.31,1.17,1.12,0.5,1.21,0.33,0.32,0.73,1.04,0.31,0.23,0.3,1.01,1,0.54,1.05,0.35,0.56,0.3,0.32,0.68,0.4,1.52,0.89,0.32,2.4,0.82,0.27,0.55,1,1.02,0.6,0.31,0.36,1.53,0.32,1.54,0.72,0.59,0.31,0.7,0.33,0.72,0.71,0.3,1.13,0.42,0.7,0.31,2.06,0.7,0.33,1.08,0.9,0.56,0.3,1.5,0.31,1.02,2.06,1.19,2,0.74,0.46,0.5,0.34,1.53,0.71,2.65,1.29,0.57,2.54,1.59,0.67,1.01,0.5,0.32,1,0.35,1.34,0.72,0.8,0.41,2.01,1.03,1.5,1,0.31,0.37,1,0.39,1.22,0.56,0.24,0.7,0.9,0.26,0.32,0.9,0.75,0.32,2.01,1.13,1.01,0.55,0.31,1.05,0.76,1,1.11,0.3,0.7,0.34,0.55,0.32,0.24,1.5,0.81,0.31,0.94,1.21,1.2,0.3,0.7,0.92,2.35,1.79,2.01,0.61,0.34,1.15,0.5,0.42,2.02,1.5,0.83,1.36,1,1,1.56,0.3,1.06,2.06,0.32,1.16,0.72,0.58,0.23,0.74,0.51,0.44,0.33,1.38,2.02,0.4,0.32,1.01,0.59,0.61,1.19,1.04,0.23,0.59,1.02,0.53,0.71,0.55,0.44,0.39,1.34,0.31,0.56,1,0.31,0.42,2.01,0.71,0.91,1.59,1.15,0.5,1.5,2.1,0.63,1.51,0.52,0.32,1.08,1.01,1.19,0.52,2.2,0.32,1.28,0.3,1.61,0.32,1.52,0.49,1.01,1.17,0.52,0.74,0.91,0.7,1,0.4,0.39,0.51,0.35,1,0.55,0.38,0.92,1.2,1.01,1.02,0.51,0.4,1.14,1.3,1,0.57,1.16,1.5,1.03,0.35,0.43,0.9,0.76,0.51,0.55,0.36,0.91,0.32,0.71,0.26,0.3,0.26,1.2,0.32,3.02,1.27,0.73,1.52,1.06,0.32,1.6,0.6,1.01,2.51,0.35,0.4,0.33,1.09,1.01,1.02,0.9,0.57,0.4,1.17,0.34,1.53,2.07,0.36,0.31,0.7,0.32,0.54,0.71,2.01,0.59,0.54,0.32,1.01,0.24,0.33,0.32,0.58,0.6,0.33,0.7,0.46,0.88,1.01,1.02,1.02,0.56,1.14,0.97,0.71,0.55,0.38,1.22,0.29,0.26,0.7,0.42,1.17,1.29,1.64,0.73,0.41,0.56,0.44,0.77,1.5,1.51,0.5,1.69,2.01,1.21,0.53,1,0.38,0.42,0.7,1.06,0.75,0.52,1.59,1.51,0.3,0.35,0.4,0.37,0.3,0.35,0.4,1,2.01,0.79,0.73,1.42,0.71,0.85,1.26,0.38,1.06,0.33,0.54,1.01,1.21,1.02,1.04,0.51,0.42,1.83,0.51,0.41,0.42,1,0.3,1.14,0.72,0.28,1.35,0.34,0.71,0.9,1.51,0.53,0.78,0.31,1.13,0.81,0.9,0.59,0.32,0.4,0.33,0.78,1.55,0.9,0.72,0.57,0.34,1.5,0.42,0.4,0.41,1.06,0.5,0.77,0.57,0.8,2.14,0.3,0.58,1.5,1.06,0.43,1.01,0.41,0.3,1.01,1.5,0.61,1.52,0.92,1.52,1.01,1.02,2.03,0.34,2.22,0.75,0.71,0.32,0.7,1.27,0.44,0.36,0.3,0.73,1.1,1.24,0.35,0.44,0.43,0.3,0.55,0.36,0.7,1,1.11,0.61,1.5,1.24,0.51,1.16,0.42,0.71,0.33,0.3,0.31,1.3,0.55,0.72,0.56,0.4,0.52,0.3,1.02,0.4,0.71,2.09,1.2,0.58,0.9,2,0.54,1.2,1.02,0.31,0.71,0.34,0.24,0.52,0.78,1,0.73,1.51,0.71,1.7,1.21,1.01,0.91,1.01,0.83,0.36,0.3,0.85,0.23,0.31,0.37,1.31,0.34,0.32,0.4,1.01,0.5,2.08,0.36,0.74,2.01,0.33,0.42,0.38,1.02,0.4,2,0.7,0.81,0.33,0.76,0.34,1.5,1.5,2.1,0.92,1.24,1.01,0.92,1.46,0.5,0.7,0.33,0.34,0.52,1.01,1.59,0.55,0.31,0.92,0.31,0.71,0.3,0.39,1.18,1.12,1.21,0.42,2.01,1.52,0.31,0.93,0.28,1.01,0.33,0.38,2,0.33,0.4,1.01,1.01,0.74,0.37,1.53,0.4,0.37,0.56,0.4,1.32,0.5,0.3,0.81,0.9,0.9,1.01,1.6,0.33,1.2,0.7,0.78,0.3,0.31,0.32,0.53,0.38,1.01,0.32,1.55,0.73,1.51,1.5,1.03,1.03,0.53,0.72,0.27,0.67,0.34,0.57,0.23,1.05,0.33,0.71,0.32,1.01,0.74,0.52,1.13,0.75,1.42,0.9,0.32,1.01,1.16,0.71,1.52,0.51,1.11,2,0.52,0.78,1,0.78,0.75,0.31,0.34,0.7,0.3,0.3,0.9,1.03,1.27,0.33,1.2,0.34,1.05,0.6,0.48,0.48,1.07,0.32,0.3,1.21,0.7,1.12,0.76,1.28,1.14,1.74,1.09,1.82,1.13,0.32,0.51,0.5,0.71,0.27,1.01,0.3,0.7,1.08,1.22,1.13,0.51,1.25,0.3,0.47,0.72,0.72,0.4,2.02,0.43,0.36,1.06,0.7,0.74,0.51,1.06,0.5,1.51,0.35,0.91,0.53,0.52,0.25,0.59,0.72,0.7,0.35,1.08,0.79,0.5,1.02,0.26,0.73,0.31,0.72,0.31,0.4,0.72,2.01,0.75,1.53,0.33,1.01,0.7,0.51,0.9,1.12,0.94,0.71,0.54,0.31,2.28,1.04,0.31,0.5,1,1,0.74,1.02,0.41,1.78,0.4,0.73,1.07,0.41,0.43,0.71,0.59,0.74,0.71,1,1.02,0.51,0.43,0.32,1.58,1.51,1.52,0.41,1.01,0.33,1.13,1.29,0.4,0.76,0.43,0.71,0.34,0.26,0.31,0.35,0.4,0.27,0.33,1.62,1.05,1.01,0.96,0.33,1.73,1.21,0.51,1.29,1.24,0.8,2.01,0.76,0.74,1,0.52,0.71,1.09,1.37,1.01,0.3,1.01,2.03,0.31,0.9,0.36,0.37,0.32,1,0.77,0.32,0.4,0.32,1.09,1.05,0.3,0.3,0.72,0.8,0.7,1.05,0.32,0.93,0.73,1.01,0.75,0.72,0.76,2.01,1.02,2.06,0.51,0.35,0.3,0.39,0.35,1.01,0.52,0.32,0.55,0.24,0.9,0.41,0.4,0.32,0.5,1,1.23,1.3,0.5,0.7,0.61,0.28,0.92,0.59,0.27,1,1.52,0.7,0.59,2.04,0.52,1.08,0.34,0.5,0.33,0.71,0.32,1.77,0.3,1.01,1.53,1.7,0.53,1.15,1.01,0.41,0.41,0.82,0.53,0.26,0.7,1,1.01,0.41,1.05,0.9,1.01,1.01,0.3,0.73,0.39,1.19,0.55,0.38,0.71,0.71,0.3,1.07,0.63,1.5,0.6,0.32,0.3,0.43,0.25,1.5,1.36,1.21,0.31,0.34,0.43,0.98,1.09,1.22,0.81,0.33,1.04,0.9,0.3,2.06,1.2,0.57,0.34,0.96,0.31,1.7,0.5,0.7,0.63,0.46,1.1,1.54,1.52,0.52,0.51,1.31,0.38,0.42,0.91,1,0.44,0.8,0.37,0.42,1.04,0.34,1.51,0.31,1.71,0.25,1.16,0.32,1.52,1.01,0.3,0.34,1.31,1.21,0.31,0.36,0.33,0.39,0.55,0.43,0.32,0.41,1.03,1.55,0.35,1.5,1.09,0.57,0.5,0.53,0.31,0.32,1.61,0.9,0.7,0.5,0.33,0.32,0.56,0.33,0.42,0.38,0.76,0.91,1.11,1,0.51,1.17,1.06,0.33,0.58,1.38,0.31,1.2,0.26,0.3,1.32,0.3,0.23,1,0.33,1.01,0.54,0.42,0.72,0.9,0.31,0.42,0.7,0.51,2.02,0.9,0.91,1.22,0.3,1.51,1.13,0.33,0.9,1.01,0.75,1.2,0.91,0.7,1.53,1.02,0.52,0.52,0.92,3.01,0.33,1.23,0.26,0.3,0.58,1.21,0.31,0.22,1.22,1.58,0.5,0.5,1.2,0.41,0.31,1.51,0.51,1.01,0.37,0.4,1.51,1.39,1.11,1.22,0.35,0.27,0.3,1,1.02,0.3,1,1.2,0.77,0.35,1.09,0.8,1.23,1.01,0.36,2.22,0.75,1.02,0.93,0.76,0.33,0.57,0.31,0.62,0.7,0.5,0.42,0.31,0.55,0.41,0.47,1.09,0.33,0.71,0.32,0.91,0.9,0.36,0.71,2.01,1.51,0.32,0.55,1.02,0.7,1.52,0.3,1.96,0.39,1.32,0.71,1.23,1,1.3,1.57,1.25,0.65,0.41,1.02,1.2,0.58,0.57,0.91,1.09,1.52,0.28,0.81,2.02,0.41,1.57,0.74,0.43,0.33,0.32,0.74,1.37,0.58,0.9,0.9,1.4,0.54,0.92,0.71,1.22,1.2,1.4,0.71,0.4,0.3,1.31,0.41,0.41,0.36,1.06,0.56,1.02,1,0.77,1.03,1.57,0.37,2.17,0.67,0.27,1.21,1.52,0.3,0.31,1.5,0.32,0.31,0.4,1.47,1.51],"line":{"colorbar":{"title":"","ticklen":2},"cmin":0.22,"cmax":3.02,"colorscale":[["0","rgba(68,1,84,1)"],["0.0416666666666667","rgba(70,19,97,1)"],["0.0833333333333333","rgba(72,32,111,1)"],["0.125","rgba(71,45,122,1)"],["0.166666666666667","rgba(68,58,128,1)"],["0.208333333333333","rgba(64,70,135,1)"],["0.25","rgba(60,82,138,1)"],["0.291666666666667","rgba(56,93,140,1)"],["0.333333333333333","rgba(49,104,142,1)"],["0.375","rgba(46,114,142,1)"],["0.416666666666667","rgba(42,123,142,1)"],["0.458333333333333","rgba(38,133,141,1)"],["0.5","rgba(37,144,140,1)"],["0.541666666666667","rgba(33,154,138,1)"],["0.583333333333333","rgba(39,164,133,1)"],["0.625","rgba(47,174,127,1)"],["0.666666666666667","rgba(53,183,121,1)"],["0.708333333333333","rgba(79,191,110,1)"],["0.75","rgba(98,199,98,1)"],["0.791666666666667","rgba(119,207,85,1)"],["0.833333333333333","rgba(147,214,70,1)"],["0.875","rgba(172,220,52,1)"],["0.916666666666667","rgba(199,225,42,1)"],["0.958333333333333","rgba(226,228,40,1)"],["1","rgba(253,231,37,1)"]],"showscale":false,"color":[0.73,0.7,0.31,0.31,0.31,0.83,0.51,0.7,0.4,1.1,0.52,0.32,1,1.01,2.01,1.6,1.01,0.31,0.38,1.03,1.31,0.7,2.01,0.8,1.2,0.31,0.82,0.41,0.33,0.33,1.3,0.35,1.25,0.62,0.3,0.4,0.9,0.4,0.5,1.09,0.91,0.34,1.66,0.42,1.17,1.27,0.9,1.55,1.5,1.06,0.9,0.42,0.32,0.38,0.78,0.3,2.01,0.34,1.31,0.9,0.33,0.31,1.22,2.15,1.27,1.06,2,2.01,0.23,1.16,1.01,0.34,0.55,0.43,0.5,0.3,1.52,1.22,0.3,0.5,1.26,0.41,0.31,0.5,0.39,0.31,1.3,1.05,1.06,0.51,0.52,1.52,0.31,1.33,0.41,0.4,0.88,0.32,0.7,0.53,0.52,0.69,0.65,1.2,1,1.2,1.5,0.38,1.51,0.35,0.3,1.01,0.54,0.31,0.5,1.04,1.19,2.13,1.1,0.3,1,0.71,1.02,0.51,0.71,0.55,0.34,0.79,0.36,0.3,0.89,0.57,1.22,1.09,0.54,1.02,0.9,1.14,0.26,0.54,0.9,1,2.02,1.5,0.53,1,2.3,1,1.51,0.62,0.81,0.33,2,0.31,1.01,0.57,0.77,1.03,1.23,1.24,0.71,0.31,1.04,1.19,0.41,0.4,1,0.53,0.27,1.11,1.63,0.9,0.42,1.73,0.41,1.3,0.24,0.35,0.3,1.01,0.27,1.16,0.59,0.7,0.5,1.5,0.55,0.36,1.05,0.3,0.9,1,0.51,0.81,0.72,0.33,0.23,0.51,0.84,0.59,0.91,0.52,0.87,0.3,0.96,0.63,1.09,0.92,1.01,0.3,0.31,1,2.2,1.01,0.71,0.72,1.31,0.77,0.51,0.59,1.54,0.57,0.71,0.51,1.31,1.01,1,1.5,1.06,0.32,0.34,0.31,1.11,0.41,0.72,0.32,0.53,0.74,0.4,0.7,1.5,1.13,0.5,0.41,1.04,0.92,1.5,0.9,0.31,1.14,0.58,1.57,0.91,0.7,0.7,0.92,0.3,2,1.32,0.58,1.05,0.7,0.35,0.7,1.5,0.58,1.01,0.31,0.32,0.7,1.52,0.7,0.7,0.92,0.9,0.34,0.33,1.5,0.42,1.05,0.31,0.7,0.32,1,1.02,0.27,1.08,0.43,1.13,1.5,0.3,0.56,1.2,0.36,0.33,0.51,2.07,1.01,0.33,0.6,0.6,0.35,0.75,1.12,0.26,1.01,0.43,0.27,1,1.03,1.01,0.33,0.24,1.01,0.3,1.02,1.5,0.5,0.3,0.71,1.11,0.28,1.29,0.57,0.33,0.75,0.35,0.32,0.32,0.29,1.03,1.7,0.9,0.32,0.61,0.51,1.13,1.02,0.3,0.31,1.51,0.81,0.7,0.74,0.51,0.35,0.62,0.32,0.58,1.01,1.07,0.7,1.04,0.51,0.53,0.3,0.9,0.78,0.33,1.51,1.02,0.43,1,0.38,0.6,0.54,0.49,0.9,1.23,1.21,0.7,0.31,0.56,1.71,0.25,0.35,0.32,0.4,0.82,0.9,0.41,0.3,1.5,0.76,0.51,0.4,0.7,0.24,0.31,0.31,0.73,0.41,0.28,1.17,1.5,1.5,1.26,1.18,1.02,0.26,0.4,1,0.35,1.01,0.34,0.73,1,0.45,0.71,2.12,0.3,0.4,0.32,0.31,0.33,1.01,1.52,1.77,1.08,0.33,0.32,0.27,0.32,1.24,0.36,0.71,0.9,1.47,0.56,1.8,2.02,0.3,0.38,0.31,0.91,0.39,0.49,0.31,0.55,0.33,0.91,0.73,0.42,1.3,1.24,0.72,1.01,1.72,0.72,1.21,0.33,0.71,0.7,0.73,1.5,1.01,2,1.5,0.71,0.34,1.29,1,0.72,0.3,1.17,0.5,0.7,0.3,0.31,2.04,1.33,0.32,1,1.52,0.71,0.3,0.33,0.34,0.34,1.01,1.52,2.02,1.64,0.4,1,0.38,0.3,1.1,1.08,1.02,0.84,1.51,2.01,0.71,0.32,0.32,1.19,1.01,0.71,0.31,0.41,0.7,1.25,0.87,1.5,0.59,1.01,0.36,0.31,0.78,0.7,0.39,0.7,1.07,0.4,1.04,0.36,0.81,0.52,0.31,1.1,0.41,1.07,0.73,0.23,0.32,0.26,0.3,0.33,0.4,1,0.73,0.71,0.38,0.39,0.41,0.35,0.55,1.01,0.42,0.31,0.43,1.12,0.51,0.52,0.3,0.44,1.01,0.9,1.56,1.01,0.33,0.3,0.9,1.7,2.02,2.17,0.28,0.53,1.59,0.5,1.07,0.53,0.42,0.33,1.08,0.32,0.44,0.73,0.84,0.41,0.32,0.31,0.3,1.51,0.5,1.04,0.34,0.31,0.41,1.51,0.91,1.22,1.2,0.51,1.51,0.81,0.31,1.24,1.01,0.36,0.7,1.04,0.5,1.02,0.31,0.41,1.12,0.42,0.27,0.68,2.14,0.53,0.7,0.31,0.7,0.34,0.41,0.57,1,0.32,0.74,0.9,0.53,1.3,1,0.5,0.95,2.01,0.51,1.11,0.75,1.5,0.72,0.74,0.7,1.01,0.24,1.01,0.43,0.43,0.41,1.24,2.03,2.01,1.23,0.78,0.4,0.63,2.32,1.12,0.56,1.59,1.04,1.01,0.32,0.7,0.42,1.5,0.32,1,2.04,1.05,1.53,1.48,1.12,1.01,0.82,1.51,0.56,0.3,0.25,0.93,1.08,0.6,1.53,1.5,0.4,0.9,1.03,1.23,0.84,0.71,0.26,0.37,0.52,2.11,1.51,1,0.55,0.51,1.01,0.32,0.58,1.26,0.78,0.84,0.23,1.5,0.33,0.51,0.73,0.52,2.04,1.54,1.53,0.24,0.55,0.52,0.44,1.19,0.35,1.76,1.01,0.41,0.76,0.5,1.09,1.21,0.52,0.39,0.5,0.92,0.37,0.7,1.71,1.21,0.52,0.51,1.2,0.31,0.41,0.81,0.72,0.83,0.63,1.8,0.32,0.34,2.14,0.7,0.3,1.21,0.54,0.41,0.43,0.32,0.24,1,0.51,0.8,1.54,0.74,1.23,0.32,0.29,1,0.41,0.41,0.55,0.57,0.39,0.4,0.82,1.5,0.7,0.34,1.03,1.03,1.04,0.61,0.31,0.72,0.98,0.29,0.27,1,0.38,1.83,0.51,1.16,0.5,1.52,0.91,1.04,1.02,1.01,0.7,0.31,0.4,0.65,0.7,0.3,1.02,0.32,0.71,0.33,0.38,0.51,0.3,0.54,1,0.3,0.31,1.51,1.02,0.31,0.76,1.12,0.71,0.55,1.12,0.58,0.41,1.02,0.71,1.05,0.33,0.7,1,0.3,1,1.5,0.86,0.26,0.35,0.5,1.5,1.01,0.32,1,0.39,0.41,1.2,1,1.01,0.73,0.31,1,0.33,0.58,0.93,1.31,1.04,0.42,1.04,1.02,0.79,0.38,1.01,1.01,0.33,0.67,0.44,1.23,0.52,0.7,0.8,0.33,1,0.37,0.74,1.2,0.31,0.71,0.51,0.76,1.27,1.7,0.31,1.53,0.83,0.52,1.06,0.53,1.51,1.51,0.71,0.34,1.52,0.9,1.54,0.56,1,1.01,0.32,1.2,0.75,0.62,1.16,0.82,0.3,1.52,0.91,0.37,0.32,1.11,1,1.01,1.5,0.58,1.06,1.5,0.32,0.91,0.51,0.31,0.38,0.27,0.9,0.78,0.55,0.44,0.7,0.54,1.04,1.04,0.7,0.43,1.04,0.32,0.8,0.38,0.5,1.51,0.5,1.19,0.94,0.45,0.32,0.4,0.25,0.79,1.52,1.22,0.43,0.31,1.08,0.43,0.7,1.21,0.3,1.22,1.21,1,0.74,1.5,1.64,0.93,0.31,0.91,0.37,1.21,0.73,0.3,2.02,1.21,0.4,1.06,0.3,1.57,0.72,0.39,0.41,0.38,0.31,0.65,1.32,0.9,0.32,1.2,0.46,0.77,1.01,1.12,1.56,0.3,0.72,1.02,1,2.22,0.53,1.22,0.56,0.34,2.24,0.33,1.05,0.31,0.73,2.09,1.09,0.7,0.57,0.73,0.33,0.3,0.33,1.73,0.51,0.31,0.57,0.8,0.71,0.32,1.25,0.34,0.32,0.68,1.51,0.52,0.3,0.52,0.32,1.23,1,0.71,0.33,1.58,0.7,1.27,0.37,0.56,1.5,1.52,0.97,0.77,0.72,0.31,0.41,1.02,1.03,1.74,1.01,0.3,0.41,0.7,1.24,1,1,0.6,1.31,0.32,0.7,0.38,1.59,1.01,0.38,0.55,2.02,0.24,0.34,0.9,0.31,0.31,1,0.97,0.4,0.62,1.03,1,0.28,0.48,1.01,1,0.43,1.01,0.38,0.3,0.7,0.91,0.66,1.01,0.32,0.33,1.56,0.79,1.01,0.71,2.01,0.5,1.04,1.21,0.41,1.16,0.32,0.58,0.38,0.34,0.5,0.32,1.15,1.05,0.54,0.3,1.29,0.4,0.33,1.03,0.7,1.2,0.3,0.33,0.5,0.6,0.62,1.07,1.13,0.71,0.32,0.91,1.13,0.9,0.71,0.3,1.69,0.58,1.59,0.28,0.53,0.39,2.16,0.31,1.55,1.6,0.4,1.01,1.03,0.92,0.62,0.3,0.4,0.7,0.9,0.82,0.5,0.92,0.31,0.91,0.4,0.4,0.26,1.11,2.48,0.32,1.09,0.42,2.01,2.02,0.7,1.01,0.32,0.3,0.7,0.38,0.31,2.09,0.41,0.3,0.9,0.7,0.28,0.9,0.54,0.28,0.31,0.93,2.02,0.41,1.37,0.37,2.11,0.23,0.51,1.09,1.18,1.17,0.67,1.6,0.76,1.04,2.01,1.06,1.24,1.06,1.16,1.13,0.51,0.5,1,1.08,1.55,0.41,0.91,0.4,1.07,0.57,1.57,0.71,0.31,0.3,0.35,0.45,0.52,0.38,1.23,0.32,0.51,0.39,0.9,1.18,1.53,0.3,0.76,0.51,0.42,0.41,1.05,0.52,1,0.28,0.23,1.01,0.77,1.51,1.7,1,0.32,1.11,0.85,1,1.08,1.5,0.51,1.5,1.26,1.01,0.31,0.53,0.41,0.53,0.43,0.51,0.8,0.71,0.34,0.9,0.5,1.51,2.01,1.64,0.33,1.04,0.78,0.76,0.52,0.44,1.11,0.9,0.56,0.79,0.71,0.73,0.4,1.03,0.32,0.52,1,1.33,1.54,1.7,1.01,0.57,1.21,0.32,1.01,1.04,0.5,0.32,1,1.3,0.31,0.5,0.43,1.24,0.32,0.53,1.15,1.21,1.04,0.27,1.04,0.71,0.27,0.32,1.5,1.01,1.02,0.56,2.06,1.2,0.73,0.32,0.73,0.9,0.25,1.01,1.09,0.41,0.31,0.53,0.52,0.58,0.38,0.73,0.35,2.04,0.32,0.75,0.5,0.72,0.5,0.32,0.53,0.72,0.79,0.35,0.7,0.59,0.33,1.45,0.3,0.32,0.31,0.45,0.32,0.31,0.7,0.96,1.5,1.01,0.34,1.2,0.43,1.04,1.13,1.22,1.09,1.29,0.4,1,0.39,0.41,0.9,1.01,0.72,1.02,1.02,0.79,2.16,0.37,0.3,0.42,0.41,0.56,1.29,0.31,0.4,0.35,1.22,0.4,0.5,1.01,0.32,0.92,0.5,0.31,0.34,0.7,2.08,0.9,0.34,0.42,0.51,0.45,2.12,0.26,0.32,0.33,0.77,0.32,0.9,1.2,1.03,0.26,0.53,1.01,0.3,0.42,0.3,1,1.01,1.51,0.33,1.31,0.39,0.51,0.9,1.01,0.39,0.32,1.12,0.53,0.5,1.5,0.32,1.45,0.78,0.3,0.4,1,2.1,1.5,1.12,1.26,1.6,0.4,0.31,0.55,1.36,0.81,1.01,2.05,0.7,0.38,1.82,0.5,0.32,1.44,0.52,1,1.07,0.8,0.72,2.03,1.38,0.4,0.74,1,2.01,1.01,1.54,0.26,0.44,0.3,0.31,0.5,0.59,2.05,1.02,0.98,0.4,0.7,0.5,0.37,0.7,0.32,0.4,0.55,0.57,0.46,0.55,0.71,1.14,0.41,1,0.3,1.1,0.5,2.01,0.4,0.34,0.3,0.34,1.51,0.45,1.08,1.13,0.3,0.7,0.34,1.14,1.07,0.43,1.2,0.5,0.41,0.31,0.57,0.93,2.03,1.01,1.07,0.73,0.52,0.53,0.37,0.71,1.12,1.51,0.32,1.87,0.41,0.7,1.09,0.38,1.24,1.2,1.14,0.62,0.7,1.01,1.01,1.1,0.5,1.53,0.31,1.01,0.55,1.5,0.32,1.22,0.31,1.11,1.13,0.53,1.03,1.06,1.04,0.72,1.05,0.3,0.7,1.59,0.34,0.73,0.41,0.81,0.73,1.21,1,1.07,0.52,0.74,0.9,1,1.03,0.41,0.39,1.27,0.33,0.34,0.33,1.22,0.52,0.71,0.31,0.8,2.06,0.51,0.31,0.5,0.32,1.01,1.02,0.34,0.77,0.32,1.21,1.47,0.72,2.42,1.01,0.72,0.33,0.7,0.7,0.4,0.51,0.5,1.59,1.2,0.61,1.01,0.7,1.13,2.02,1.23,0.31,0.71,0.3,1.17,0.32,0.92,0.32,2.19,0.53,0.3,0.77,0.32,0.32,0.3,0.96,0.42,0.35,0.31,0.81,0.53,1.2,2.01,0.41,1.54,0.95,1.61,1.04,0.34,0.3,0.51,0.8,1.17,1,0.51,0.32,0.41,1.1,2.39,1.7,1.28,1,0.52,2.04,1.01,1.01,1.5,0.53,2.01,0.71,0.31,0.34,0.42,0.36,0.3,0.41,0.6,0.33,0.35,0.36,1,1.05,0.41,0.32,0.56,1.18,0.49,1.05,1.11,0.52,1.14,0.36,0.52,1.35,0.5,0.91,0.52,0.62,1.51,0.71,0.4,0.48,0.91,1.57,0.55,1.51,0.4,1.24,0.3,0.32,0.3,0.7,0.34,0.22,1.56,0.53,1.01,0.56,1.25,1.01,0.79,0.43,0.5,1.01,1.59,1.2,0.28,1.09,0.76,0.9,0.4,0.54,1.51,0.77,0.79,0.5,0.23,0.78,0.63,0.91,0.36,0.33,0.31,0.38,0.38,1.21,0.33,2.03,1.01,0.33,2.18,1.54,0.41,0.33,0.7,1.12,0.51,1.16,0.91,1.5,0.3,2.01,0.33,0.55,0.3,0.4,0.71,1.5,0.91,0.5,0.7,0.52,0.39,0.28,1.58,1.34,0.91,0.41,0.33,0.56,1.05,1.03,0.53,0.93,0.31,1,1.26,0.42,1.06,0.38,1.41,0.4,1.51,1.52,1.08,1.5,0.44,0.3,1.27,0.3,1.1,1.54,0.9,0.4,0.35,1.09,0.71,0.57,1.5,2.09,0.3,2.01,0.5,0.41,0.82,0.42,0.7,0.31,0.59,0.55,1.5,1.03,1.01,0.35,0.56,0.7,0.9,0.38,0.72,1.13,0.61,0.32,0.78,1.03,0.23,1.01,0.71,0.37,1.21,0.5,0.53,2.21,0.7,0.45,0.66,0.33,0.52,0.3,2.04,0.36,0.34,1.01,1.5,0.24,0.34,0.71,1.07,1,0.27,2.14,2.24,0.7,1.19,1.5,0.56,0.79,0.77,0.34,1.01,0.3,0.71,1.01,0.34,0.7,0.3,1.03,0.57,1.13,0.71,0.62,0.41,0.3,0.71,0.92,1.24,0.53,1.01,0.4,0.7,1.02,0.59,0.72,2.03,0.57,0.35,1.06,1.21,0.76,0.42,0.4,1.1,0.77,1.2,1.34,0.6,0.77,1.52,1.23,0.72,0.3,0.42,1.52,2.11,0.58,0.36,0.51,0.93,0.64,0.94,1.7,0.25,0.32,0.56,0.95,0.29,0.33,0.41,1,1.02,0.3,2.05,0.7,0.55,1.58,1.01,2.03,0.5,0.32,0.38,1.08,0.38,0.59,0.9,1.03,0.78,1.5,1.54,2.05,0.53,1.24,0.45,0.51,0.25,0.36,0.58,0.51,0.9,0.7,0.3,0.4,0.41,0.31,0.53,0.25,0.55,0.41,0.7,0.73,0.32,0.5,0.33,0.31,1.51,0.8,1.41,0.34,0.41,0.7,0.39,0.29,2.38,0.9,0.36,0.7,0.25,1.01,1.16,1.01,0.7,0.3,0.9,0.4,1.6,0.3,0.9,0.7,0.51,0.3,1,0.55,0.5,0.31,0.57,1.01,1.02,0.5,0.25,0.72,1.23,1.26,0.52,0.7,0.32,1.28,0.7,0.53,0.3,1.88,0.9,1.52,1.4,0.32,1.04,0.9,2.1,2,0.36,0.33,0.32,0.54,0.4,1.09,1.01,1.03,0.78,0.42,0.34,1.06,0.32,0.71,0.31,0.37,0.34,0.7,0.35,0.43,0.5,1,0.9,0.28,0.41,1.18,1.02,1.2,0.74,1.2,0.7,0.53,0.26,0.32,1.15,1.07,0.52,1.04,1.01,0.7,0.56,1.01,0.32,1.11,1,0.51,1.7,1.43,0.55,0.51,1.51,0.74,0.9,0.37,1.01,1.04,0.31,1.01,2.07,0.31,0.31,0.8,0.72,0.82,0.31,0.39,2.08,0.42,0.34,0.36,0.6,0.9,0.74,0.74,1.48,0.33,0.37,0.71,0.95,0.73,0.37,1.5,1.13,0.4,1,0.85,0.51,1.01,1.02,1.54,0.92,0.76,1.04,0.9,0.37,1.02,1,0.3,0.36,0.41,0.71,0.5,0.34,1.74,0.3,0.4,1.2,1.21,0.57,1.12,0.58,0.7,1.07,0.7,0.7,1.7,1.01,0.33,0.52,0.27,0.4,0.42,0.25,0.37,1.14,0.3,0.34,0.36,0.31,0.91,1.17,0.34,0.33,0.3,0.42,0.3,1.01,0.4,0.41,0.3,0.32,1.02,2.01,0.3,0.31,0.7,1.02,1.04,1.1,1.24,0.61,2.14,1.02,0.31,1.15,0.4,0.26,0.34,0.8,1.21,0.33,0.31,1.51,1.34,0.53,1.17,0.3,2,0.3,2.1,0.32,0.5,0.53,0.31,0.8,0.32,2.01,1.11,0.36,0.72,1.25,1.2,0.73,1.07,0.51,0.58,0.73,1.5,0.53,0.7,0.43,1.5,0.52,1.25,2.1,0.31,0.6,1.01,0.31,0.45,0.83,0.5,0.41,2.5,2,0.7,1.13,0.31,1.01,0.83,0.41,0.5,1.51,1.3,0.71,0.3,0.71,0.46,1.03,0.37,2.51,1.07,1,0.7,0.25,0.34,0.3,0.55,0.4,0.51,0.3,0.51,0.42,0.51,1.11,0.4,0.43,0.36,1.16,0.52,1.03,0.33,0.41,0.53,0.27,0.38,0.34,0.41,0.3,0.3,0.46,1.56,0.41,0.8,0.24,1.86,0.57,0.28,1.04,0.32,0.31,1.1,0.9,0.57,0.3,0.66,0.32,0.52,1.34,0.3,1.22,0.3,2.51,1.52,1.01,1,0.34,0.52,0.46,0.52,0.96,0.4,1.62,0.32,1.07,1.74,0.35,1.74,1.01,1.01,0.71,0.75,0.32,0.74,0.71,1.01,2.08,0.34,0.75,1.01,0.31,1.24,0.31,0.69,1.24,0.53,0.24,1.01,1.3,1.09,0.75,0.32,0.7,1.21,0.3,0.71,0.38,0.5,2.02,1.19,0.25,0.33,0.41,0.34,0.34,0.3,0.41,0.93,0.45,0.71,0.38,1.57,0.32,1.55,0.9,1.2,0.76,0.6,0.95,0.73,2.3,0.3,0.3,1.54,0.51,0.55,0.31,0.26,0.9,0.5,1.01,0.93,0.5,0.41,2.01,1.1,0.54,0.41,1.2,0.34,1.7,0.77,1.11,0.9,1.02,2.6,0.83,1.15,0.3,0.34,1.65,1.51,1.26,0.37,0.72,0.47,0.74,0.7,0.31,0.71,0.4,0.3,0.54,0.3,0.32,0.36,0.54,0.32,1.09,1.18,0.35,1.29,0.3,0.41,0.3,0.7,0.37,0.24,0.41,2.04,0.32,0.71,0.3,0.71,0.51,0.56,0.27,0.27,1.01,2.01,0.3,0.54,0.39,0.38,0.91,1.01,0.92,0.62,2.44,1,0.38,0.57,1,1,0.51,0.31,0.94,0.76,1,0.93,0.84,0.34,0.72,0.31,0.23,1.16,0.5,0.5,1.66,1.01,1.58,0.57,1.56,2,0.7,0.4,1.02,0.73,0.32,0.55,1.03,0.41,0.33,1.35,1.51,0.53,1.06,1.2,1.25,1.01,0.51,0.7,0.71,1.03,0.31,0.36,0.36,0.32,0.3,1.29,0.31,1,1.04,2.02,0.42,0.53,0.3,0.53,0.55,0.3,0.3,0.29,0.62,1.05,1.51,0.32,0.62,0.37,0.32,0.3,0.42,0.57,1.51,0.98,0.56,1.27,0.3,1,0.31,0.32,1.27,0.55,0.31,1.16,0.39,0.41,0.42,2,0.55,1.59,0.31,0.9,0.43,0.31,0.9,0.42,0.85,0.23,0.8,0.32,1.01,0.59,0.7,0.27,0.64,1.08,0.26,0.43,0.33,1.23,1.51,0.94,1.23,0.3,0.31,2.19,0.38,0.9,0.34,0.35,0.33,1.51,0.32,0.5,1.01,0.91,1.51,1,1.13,0.5,0.8,0.36,0.36,1.25,1.58,0.32,0.3,0.8,0.39,0.42,0.42,0.7,0.54,0.42,0.74,1.02,0.31,0.36,2.02,0.91,1.57,0.33,1.05,1.01,0.51,1.02,0.7,1.18,1.12,0.32,2.13,0.3,1.02,0.38,0.72,2.01,0.41,1.25,1.04,1.18,1,0.72,1.2,0.55,1,1.05,1.51,0.31,0.75,0.56,1.01,0.41,0.7,1.01,0.71,1.2,1,1.51,0.9,1.01,0.43,0.46,1.5,0.41,0.42,1.51,0.5,1.02,0.75,0.53,0.8,0.7,1.01,0.9,0.61,0.55,0.31,0.52,0.31,0.74,0.41,0.31,0.56,0.36,0.5,1.8,0.71,0.52,1.01,0.53,2,2.16,0.53,0.32,0.36,0.84,0.7,0.31,0.7,0.4,1.02,1,1.33,1,1.08,0.53,0.3,0.5,1.01,0.8,0.54,0.81,0.56,1.01,0.32,0.54,0.3,0.3,0.4,1.01,1.17,0.35,0.32,1.51,0.4,1.6,0.31,0.31,0.99,2.01,0.27,0.3,0.7,0.67,0.24,0.71,0.54,0.71,2.09,0.32,1.01,1.06,0.7,0.3,2.1,0.51,0.54,1.03,1.51,0.34,0.41,0.46,1.04,0.33,0.51,0.34,0.31,1.23,0.33,0.34,0.31,0.65,1.58,0.23,2.02,1.83,0.91,0.31,1.02,1,0.36,1.51,1.34,0.92,1.16,0.3,1.01,2.15,0.9,0.9,1.33,0.54,0.33,0.31,1.2,0.41,0.51,1.85,0.32,1.32,0.76,0.23,0.54,0.32,1.02,0.5,1.02,0.44,1.01,1.23,0.9,1.53,1.3,0.73,0.31,0.34,0.27,1.63,0.5,0.74,1.16,0.3,2,0.3,1.32,0.56,0.38,0.32,1.12,1.2,0.53,0.37,1.61,0.79,1.51,1.04,1.01,1.53,0.3,0.3,1.03,0.3,1.05,0.31,0.3,0.31,0.36,0.51,0.75,0.71,0.38,0.43,1.06,0.72,1.25,0.9,0.53,1.21,0.83,1.52,0.3,1.5,1.21,0.77,0.39,0.4,0.32,0.92,0.36,1.02,0.25,0.3,0.3,1.5,1.01,1.01,1.06,0.9,0.7,0.5,1.59,1.18,1.11,1.04,0.74,0.28,1.08,0.52,0.36,1.03,0.36,2,0.7,0.77,0.7,0.31,0.35,0.42,0.67,0.56,2,0.9,0.42,0.33,0.6,0.41,1.01,0.83,0.3,0.34,0.35,0.76,0.9,2,1.51,0.7,0.3,0.7,1.01,1.13,1.52,0.4,0.31,1.09,0.52,0.39,1.39,0.41,0.7,1.01,1.1,0.32,0.35,0.31,0.3,1.52,0.9,0.37,0.71,1.64,0.59,1.09,2.01,0.41,1.26,0.76,0.35,0.7,1.51,0.28,0.9,0.96,1.03,1.51,1.1,1.09,1.03,1.06,0.31,1.53,0.52,0.7,1.5,1,1.07,0.31,0.33,0.4,0.32,0.91,0.5,0.32,0.37,1.1,0.34,0.51,0.4,1.14,1.05,0.32,1,0.51,1.02,0.32,0.32,0.3,0.71,1.01,0.39,1.52,0.44,0.54,0.72,1.01,1.04,0.9,0.3,1.7,1.01,0.93,0.5,0.25,0.3,0.71,0.41,1.31,1.16,0.71,0.3,1,1.13,0.34,0.68,0.4,0.53,0.7,0.41,1.61,1.21,0.32,0.63,1.13,0.71,0.23,1.01,0.81,1,1.22,1.53,0.32,0.33,0.39,1.08,0.92,0.72,0.54,0.73,0.36,0.71,0.7,0.34,0.41,0.72,0.31,1,1.77,0.9,2.01,0.54,1.21,1.7,0.44,0.9,1.22,1.55,0.81,0.5,1.3,0.7,1.02,0.5,0.33,0.41,0.76,0.61,1.2,0.58,0.27,1.2,1.01,0.51,0.31,0.4,0.71,1.56,0.41,0.4,2.36,1.55,0.24,0.51,1.1,1.02,1,0.5,1.5,0.73,0.31,0.56,0.38,0.54,0.38,0.77,0.51,0.73,0.71,0.42,1.51,0.52,1.55,0.76,1.01,0.73,0.91,0.39,0.33,0.37,1.01,0.41,1.12,1.15,0.42,0.42,0.3,0.38,1.2,0.42,0.7,0.71,2,0.48,0.29,0.38,0.9,1.3,0.33,2.52,0.33,0.62,0.31,0.54,1.04,0.4,0.42,0.51,0.92,0.5,0.41,0.7,0.7,1.59,0.3,1.51,1.04,0.31,1.05,1,2.01,0.55,0.71,0.23,1.5,0.71,0.71,2,0.7,0.79,2.28,0.37,1.27,0.36,1,0.82,0.42,0.57,1.26,1.39,1,1.01,1.15,0.33,0.24,0.7,1.34,0.37,0.31,2.2,1.08,1,0.52,1.01,1,0.75,0.41,0.24,1.5,1.51,0.8,1,0.54,1.07,1.08,1.5,0.41,1.5,0.7,0.37,0.8,1.02,0.5,0.78,0.31,1.06,0.5,1.15,0.25,2.26,1.05,0.57,1.01,0.32,0.53,0.91,0.41,1.23,1.31,0.9,0.42,1.13,0.34,1.05,0.58,0.3,1,0.42,0.91,0.32,0.86,1.33,1.52,0.72,0.7,0.53,0.5,0.42,1.03,1.16,2.04,0.4,1.01,0.5,1.04,0.75,1.26,1.06,0.34,0.32,1.01,0.91,0.36,0.9,1.2,0.9,1.23,0.75,0.5,0.53,0.3,0.91,0.51,2,0.75,0.9,1.42,0.34,1.3,1.51,1.01,1.27,2.01,0.4,0.27,1.03,0.73,2.02,0.38,0.36,1.51,1,1.51,0.32,1.09,1.34,0.4,1.04,0.7,0.3,0.31,0.76,0.27,0.33,0.64,0.39,0.7,1.02,1,0.72,0.7,1.23,0.74,0.3,0.42,0.71,0.76,0.38,0.9,1.02,1.67,0.51,0.9,1.2,0.9,1.53,1.11,0.41,1.32,0.38,0.35,1.52,0.9,0.24,0.75,0.91,0.4,0.33,1.31,0.36,2.01,0.33,1.51,1.25,1.02,1.22,0.34,0.33,0.7,0.77,0.5,0.31,0.3,0.56,2.02,1.02,1.01,1.51,2.04,0.71,1.57,0.35,0.53,0.32,0.78,0.53,2.1,0.82,0.71,0.3,1.05,0.7,0.33,2.01,2.01,1.01,0.42,1.02,0.93,0.81,2.61,1.18,0.7,0.28,0.39,0.71,1.59,1.04,0.53,0.9,0.32,1.18,0.31,0.31,0.3,0.4,1.54,1.11,0.57,1.5,1.65,0.72,0.29,0.52,0.76,1.06,1.62,0.71,1.21,0.52,1.12,0.96,1.01,0.3,0.32,1.51,0.3,1.16,0.66,0.79,0.59,0.57,1.01,1.26,0.44,0.5,1.5,0.51,1.2,0.71,0.9,1.06,1.24,0.53,0.72,1.01,0.36,1.14,0.3,1.58,1.11,1.21,0.3,0.33,0.81,0.39,0.51,1.01,0.42,0.39,0.92,1,1.57,0.32,0.91,0.44,0.53,0.58,0.32,0.7,0.38,0.3,0.7,0.77,2.01,0.71,1.71,0.42,2.06,1.98,1.01,0.54,0.5,1.04,0.36,0.61,0.24,0.91,1,0.31,0.31,1.73,0.3,0.31,0.42,1.01,0.52,1,1.27,1.22,0.54,0.31,0.98,0.84,0.31,0.33,0.3,2.5,1.52,0.38,0.75,0.43,1.02,0.77,0.35,0.33,0.5,1.02,0.72,0.41,0.38,0.7,0.41,1.01,0.31,1.08,2.02,1.56,0.73,1.01,0.8,0.35,0.71,0.41,0.86,0.9,1.46,0.4,1.22,0.53,0.71,1.52,0.3,0.42,0.42,1.41,0.3,1.51,0.5,1.68,1.53,0.54,1.14,2.4,0.42,1.03,0.9,1.05,1,0.41,1,0.7,0.3,0.54,1.01,0.71,0.7,1.01,0.4,0.58,0.38,1.08,0.32,0.34,0.3,0.32,1.51,0.33,0.71,0.48,1.09,0.72,1.67,0.75,0.27,0.41,0.55,0.46,0.34,0.7,0.54,1.01,2,0.4,1.07,0.51,0.51,1.51,1.01,1.02,0.31,0.3,0.3,1,0.36,0.51,1.02,0.7,0.58,0.33,1.01,1,0.76,0.3,2.01,0.9,0.35,0.52,0.34,1.03,1.03,0.29,0.36,0.42,0.3,0.55,0.5,1,0.31,0.72,0.54,0.3,1.56,0.9,1.11,0.83,0.28,0.43,0.55,0.33,1.02,1.01,0.57,1.01,0.55,0.33,0.9,0.33,0.54,0.41,1.15,1.03,0.41,0.59,0.31,1.62,0.3,1.37,1.01,0.31,0.32,0.58,1.54,1.13,0.8,0.4,0.96,0.56,1,0.5,0.3,0.51,1.5,1.01,0.58,0.53,0.38,0.56,0.28,0.53,0.86,0.59,0.27,0.73,0.6,0.35,0.57,1.5,1.13,1.51,0.81,0.23,0.3,0.78,0.25,1.5,0.93,0.52,1.23,0.73,0.32,0.72,1.5,1.12,1.5,0.42,0.3,0.34,0.92,0.43,1.1,0.87,0.96,0.71,1.23,0.24,0.6,0.7,0.3,1.2,0.34,1.6,1.52,1.09,1.2,1.59,0.24,0.41,0.54,1.31,0.32,0.35,0.31,1,1.95,0.32,0.4,0.71,0.7,1.15,1.21,0.52,0.7,0.82,0.26,0.92,1.11,0.43,2.2,1.38,0.51,1.01,1.01,2.03,0.52,1.21,0.44,1,1.04,1.29,0.41,1.08,0.71,1.65,0.52,1.11,0.32,0.74,0.3,0.3,0.41,0.63,1,0.7,1.16,0.4,0.69,1.01,0.33,0.5,1.79,1.05,1.09,0.79,0.51,0.35,1.02,1.06,1.01,0.5,1.51,0.3,0.31,1.2,1.03,0.41,1.11,0.33,0.54,0.32,1.26,0.46,0.44,0.33,0.58,0.3,0.5,1.15,0.76,1.5,0.51,2,0.72,1.12,0.7,0.7,0.32,1.04,0.81,0.44,0.38,0.32,2.03,0.39,1.34,0.77,0.51,0.33,0.9,1.53,1,0.38,1,1,1.7,0.55,1.13,0.57,0.31,0.41,0.9,0.32,0.55,0.91,1.5,0.33,0.35,1.5,1.01,0.3,0.34,0.33,1.01,1.04,0.41,0.71,1.05,1.07,2.02,0.34,0.73,0.71,1.24,0.26,0.31,0.38,0.34,1.2,0.5,0.42,1.75,0.61,0.36,0.53,0.9,1.36,0.24,1.23,0.31,1.01,0.36,0.3,1.05,0.32,2.02,1.21,0.65,0.7,0.41,0.43,0.41,0.4,0.55,0.52,1.02,1.02,0.35,0.31,1.03,0.72,0.41,0.71,0.47,0.31,2.01,0.5,0.34,1.23,1.14,1.53,1.03,1.71,1.33,1.04,0.3,0.33,1,0.3,1.02,0.54,0.83,1.05,0.31,0.32,2.04,0.73,1.01,1,0.3,0.44,0.53,1.71,1.02,0.61,0.71,1.41,1.11,0.52,0.74,0.9,0.31,0.32,1,0.7,1,0.72,0.4,1.59,0.83,1.61,0.3,1.26,0.56,0.38,1.12,0.74,0.31,0.42,0.83,0.5,0.81,0.71,0.31,0.7,0.53,0.33,0.31,1.17,1.12,0.5,1.21,0.33,0.32,0.73,1.04,0.31,0.23,0.3,1.01,1,0.54,1.05,0.35,0.56,0.3,0.32,0.68,0.4,1.52,0.89,0.32,2.4,0.82,0.27,0.55,1,1.02,0.6,0.31,0.36,1.53,0.32,1.54,0.72,0.59,0.31,0.7,0.33,0.72,0.71,0.3,1.13,0.42,0.7,0.31,2.06,0.7,0.33,1.08,0.9,0.56,0.3,1.5,0.31,1.02,2.06,1.19,2,0.74,0.46,0.5,0.34,1.53,0.71,2.65,1.29,0.57,2.54,1.59,0.67,1.01,0.5,0.32,1,0.35,1.34,0.72,0.8,0.41,2.01,1.03,1.5,1,0.31,0.37,1,0.39,1.22,0.56,0.24,0.7,0.9,0.26,0.32,0.9,0.75,0.32,2.01,1.13,1.01,0.55,0.31,1.05,0.76,1,1.11,0.3,0.7,0.34,0.55,0.32,0.24,1.5,0.81,0.31,0.94,1.21,1.2,0.3,0.7,0.92,2.35,1.79,2.01,0.61,0.34,1.15,0.5,0.42,2.02,1.5,0.83,1.36,1,1,1.56,0.3,1.06,2.06,0.32,1.16,0.72,0.58,0.23,0.74,0.51,0.44,0.33,1.38,2.02,0.4,0.32,1.01,0.59,0.61,1.19,1.04,0.23,0.59,1.02,0.53,0.71,0.55,0.44,0.39,1.34,0.31,0.56,1,0.31,0.42,2.01,0.71,0.91,1.59,1.15,0.5,1.5,2.1,0.63,1.51,0.52,0.32,1.08,1.01,1.19,0.52,2.2,0.32,1.28,0.3,1.61,0.32,1.52,0.49,1.01,1.17,0.52,0.74,0.91,0.7,1,0.4,0.39,0.51,0.35,1,0.55,0.38,0.92,1.2,1.01,1.02,0.51,0.4,1.14,1.3,1,0.57,1.16,1.5,1.03,0.35,0.43,0.9,0.76,0.51,0.55,0.36,0.91,0.32,0.71,0.26,0.3,0.26,1.2,0.32,3.02,1.27,0.73,1.52,1.06,0.32,1.6,0.6,1.01,2.51,0.35,0.4,0.33,1.09,1.01,1.02,0.9,0.57,0.4,1.17,0.34,1.53,2.07,0.36,0.31,0.7,0.32,0.54,0.71,2.01,0.59,0.54,0.32,1.01,0.24,0.33,0.32,0.58,0.6,0.33,0.7,0.46,0.88,1.01,1.02,1.02,0.56,1.14,0.97,0.71,0.55,0.38,1.22,0.29,0.26,0.7,0.42,1.17,1.29,1.64,0.73,0.41,0.56,0.44,0.77,1.5,1.51,0.5,1.69,2.01,1.21,0.53,1,0.38,0.42,0.7,1.06,0.75,0.52,1.59,1.51,0.3,0.35,0.4,0.37,0.3,0.35,0.4,1,2.01,0.79,0.73,1.42,0.71,0.85,1.26,0.38,1.06,0.33,0.54,1.01,1.21,1.02,1.04,0.51,0.42,1.83,0.51,0.41,0.42,1,0.3,1.14,0.72,0.28,1.35,0.34,0.71,0.9,1.51,0.53,0.78,0.31,1.13,0.81,0.9,0.59,0.32,0.4,0.33,0.78,1.55,0.9,0.72,0.57,0.34,1.5,0.42,0.4,0.41,1.06,0.5,0.77,0.57,0.8,2.14,0.3,0.58,1.5,1.06,0.43,1.01,0.41,0.3,1.01,1.5,0.61,1.52,0.92,1.52,1.01,1.02,2.03,0.34,2.22,0.75,0.71,0.32,0.7,1.27,0.44,0.36,0.3,0.73,1.1,1.24,0.35,0.44,0.43,0.3,0.55,0.36,0.7,1,1.11,0.61,1.5,1.24,0.51,1.16,0.42,0.71,0.33,0.3,0.31,1.3,0.55,0.72,0.56,0.4,0.52,0.3,1.02,0.4,0.71,2.09,1.2,0.58,0.9,2,0.54,1.2,1.02,0.31,0.71,0.34,0.24,0.52,0.78,1,0.73,1.51,0.71,1.7,1.21,1.01,0.91,1.01,0.83,0.36,0.3,0.85,0.23,0.31,0.37,1.31,0.34,0.32,0.4,1.01,0.5,2.08,0.36,0.74,2.01,0.33,0.42,0.38,1.02,0.4,2,0.7,0.81,0.33,0.76,0.34,1.5,1.5,2.1,0.92,1.24,1.01,0.92,1.46,0.5,0.7,0.33,0.34,0.52,1.01,1.59,0.55,0.31,0.92,0.31,0.71,0.3,0.39,1.18,1.12,1.21,0.42,2.01,1.52,0.31,0.93,0.28,1.01,0.33,0.38,2,0.33,0.4,1.01,1.01,0.74,0.37,1.53,0.4,0.37,0.56,0.4,1.32,0.5,0.3,0.81,0.9,0.9,1.01,1.6,0.33,1.2,0.7,0.78,0.3,0.31,0.32,0.53,0.38,1.01,0.32,1.55,0.73,1.51,1.5,1.03,1.03,0.53,0.72,0.27,0.67,0.34,0.57,0.23,1.05,0.33,0.71,0.32,1.01,0.74,0.52,1.13,0.75,1.42,0.9,0.32,1.01,1.16,0.71,1.52,0.51,1.11,2,0.52,0.78,1,0.78,0.75,0.31,0.34,0.7,0.3,0.3,0.9,1.03,1.27,0.33,1.2,0.34,1.05,0.6,0.48,0.48,1.07,0.32,0.3,1.21,0.7,1.12,0.76,1.28,1.14,1.74,1.09,1.82,1.13,0.32,0.51,0.5,0.71,0.27,1.01,0.3,0.7,1.08,1.22,1.13,0.51,1.25,0.3,0.47,0.72,0.72,0.4,2.02,0.43,0.36,1.06,0.7,0.74,0.51,1.06,0.5,1.51,0.35,0.91,0.53,0.52,0.25,0.59,0.72,0.7,0.35,1.08,0.79,0.5,1.02,0.26,0.73,0.31,0.72,0.31,0.4,0.72,2.01,0.75,1.53,0.33,1.01,0.7,0.51,0.9,1.12,0.94,0.71,0.54,0.31,2.28,1.04,0.31,0.5,1,1,0.74,1.02,0.41,1.78,0.4,0.73,1.07,0.41,0.43,0.71,0.59,0.74,0.71,1,1.02,0.51,0.43,0.32,1.58,1.51,1.52,0.41,1.01,0.33,1.13,1.29,0.4,0.76,0.43,0.71,0.34,0.26,0.31,0.35,0.4,0.27,0.33,1.62,1.05,1.01,0.96,0.33,1.73,1.21,0.51,1.29,1.24,0.8,2.01,0.76,0.74,1,0.52,0.71,1.09,1.37,1.01,0.3,1.01,2.03,0.31,0.9,0.36,0.37,0.32,1,0.77,0.32,0.4,0.32,1.09,1.05,0.3,0.3,0.72,0.8,0.7,1.05,0.32,0.93,0.73,1.01,0.75,0.72,0.76,2.01,1.02,2.06,0.51,0.35,0.3,0.39,0.35,1.01,0.52,0.32,0.55,0.24,0.9,0.41,0.4,0.32,0.5,1,1.23,1.3,0.5,0.7,0.61,0.28,0.92,0.59,0.27,1,1.52,0.7,0.59,2.04,0.52,1.08,0.34,0.5,0.33,0.71,0.32,1.77,0.3,1.01,1.53,1.7,0.53,1.15,1.01,0.41,0.41,0.82,0.53,0.26,0.7,1,1.01,0.41,1.05,0.9,1.01,1.01,0.3,0.73,0.39,1.19,0.55,0.38,0.71,0.71,0.3,1.07,0.63,1.5,0.6,0.32,0.3,0.43,0.25,1.5,1.36,1.21,0.31,0.34,0.43,0.98,1.09,1.22,0.81,0.33,1.04,0.9,0.3,2.06,1.2,0.57,0.34,0.96,0.31,1.7,0.5,0.7,0.63,0.46,1.1,1.54,1.52,0.52,0.51,1.31,0.38,0.42,0.91,1,0.44,0.8,0.37,0.42,1.04,0.34,1.51,0.31,1.71,0.25,1.16,0.32,1.52,1.01,0.3,0.34,1.31,1.21,0.31,0.36,0.33,0.39,0.55,0.43,0.32,0.41,1.03,1.55,0.35,1.5,1.09,0.57,0.5,0.53,0.31,0.32,1.61,0.9,0.7,0.5,0.33,0.32,0.56,0.33,0.42,0.38,0.76,0.91,1.11,1,0.51,1.17,1.06,0.33,0.58,1.38,0.31,1.2,0.26,0.3,1.32,0.3,0.23,1,0.33,1.01,0.54,0.42,0.72,0.9,0.31,0.42,0.7,0.51,2.02,0.9,0.91,1.22,0.3,1.51,1.13,0.33,0.9,1.01,0.75,1.2,0.91,0.7,1.53,1.02,0.52,0.52,0.92,3.01,0.33,1.23,0.26,0.3,0.58,1.21,0.31,0.22,1.22,1.58,0.5,0.5,1.2,0.41,0.31,1.51,0.51,1.01,0.37,0.4,1.51,1.39,1.11,1.22,0.35,0.27,0.3,1,1.02,0.3,1,1.2,0.77,0.35,1.09,0.8,1.23,1.01,0.36,2.22,0.75,1.02,0.93,0.76,0.33,0.57,0.31,0.62,0.7,0.5,0.42,0.31,0.55,0.41,0.47,1.09,0.33,0.71,0.32,0.91,0.9,0.36,0.71,2.01,1.51,0.32,0.55,1.02,0.7,1.52,0.3,1.96,0.39,1.32,0.71,1.23,1,1.3,1.57,1.25,0.65,0.41,1.02,1.2,0.58,0.57,0.91,1.09,1.52,0.28,0.81,2.02,0.41,1.57,0.74,0.43,0.33,0.32,0.74,1.37,0.58,0.9,0.9,1.4,0.54,0.92,0.71,1.22,1.2,1.4,0.71,0.4,0.3,1.31,0.41,0.41,0.36,1.06,0.56,1.02,1,0.77,1.03,1.57,0.37,2.17,0.67,0.27,1.21,1.52,0.3,0.31,1.5,0.32,0.31,0.4,1.47,1.51]}},"xaxis":"x","yaxis":"y","frame":null},{"x":[0.22,3.02],"y":[335,18795],"type":"scatter","mode":"markers","opacity":0,"hoverinfo":"none","showlegend":false,"marker":{"colorbar":{"title":"carat","ticklen":2,"len":0.5,"lenmode":"fraction","y":1,"yanchor":"top"},"cmin":0.22,"cmax":3.02,"colorscale":[["0","rgba(68,1,84,1)"],["0.0416666666666667","rgba(70,19,97,1)"],["0.0833333333333333","rgba(72,32,111,1)"],["0.125","rgba(71,45,122,1)"],["0.166666666666667","rgba(68,58,128,1)"],["0.208333333333333","rgba(64,70,135,1)"],["0.25","rgba(60,82,138,1)"],["0.291666666666667","rgba(56,93,140,1)"],["0.333333333333333","rgba(49,104,142,1)"],["0.375","rgba(46,114,142,1)"],["0.416666666666667","rgba(42,123,142,1)"],["0.458333333333333","rgba(38,133,141,1)"],["0.5","rgba(37,144,140,1)"],["0.541666666666667","rgba(33,154,138,1)"],["0.583333333333333","rgba(39,164,133,1)"],["0.625","rgba(47,174,127,1)"],["0.666666666666667","rgba(53,183,121,1)"],["0.708333333333333","rgba(79,191,110,1)"],["0.75","rgba(98,199,98,1)"],["0.791666666666667","rgba(119,207,85,1)"],["0.833333333333333","rgba(147,214,70,1)"],["0.875","rgba(172,220,52,1)"],["0.916666666666667","rgba(199,225,42,1)"],["0.958333333333333","rgba(226,228,40,1)"],["1","rgba(253,231,37,1)"]],"showscale":true,"color":[0.22,3.02],"line":{"color":"rgba(255,127,14,1)"}},"xaxis":"x","yaxis":"y","frame":null}],"highlight":{"on":"plotly_click","persistent":false,"dynamic":false,"selectize":false,"opacityDim":0.2,"selected":{"opacity":1},"debounce":0},"shinyEvents":["plotly_hover","plotly_click","plotly_selected","plotly_relayout","plotly_brushed","plotly_brushing","plotly_clickannotation","plotly_doubleclick","plotly_deselect","plotly_afterplot","plotly_sunburstclick"],"base_url":"https://plot.ly"},"evals":[],"jsHooks":[]}</script> --- # **plotly** with **ggplot2** style Use function `ggplotly()` on a **ggplot2** syntax. Example: ```r p <- ggplot(diam_ech) + aes(x = color, y = price) + geom_boxplot() ggplotly(p) ``` <div class="plotly html-widget html-fill-item-overflow-hidden html-fill-item" id="htmlwidget-511dd718563422479768" style="width:360px;height:216px;"></div> <script type="application/json" data-for="htmlwidget-511dd718563422479768">{"x":{"data":[{"x":[6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,6,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,4,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,1,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,5,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,2,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,3,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7,7],"y":[2397,1669,1378,13753,10147,1259,5291,4078,8732,4640,13500,14027,11086,10462,17422,4749,4525,7891,3360,4326,5546,18756,17153,1173,974,5672,6694,18255,13669,403,1427,8475,4189,2927,628,938,2700,4879,4466,449,12883,4155,4560,8371,8521,2175,3604,4584,540,5285,5458,2089,3849,534,532,589,6303,1883,1211,978,13512,7715,635,14388,13109,14959,4771,4620,9439,5699,2428,16041,5412,16874,9209,2120,10824,803,884,2239,6383,5557,4657,12451,4844,2386,3276,1698,2761,16309,1140,651,15189,2557,8896,1917,5960,2255,1626,449,1232,3605,6206,15654,1180,11954,4355,480,771,6887,10577,4791,1046,2642,5352,1715,2609,1052,6756,2329,5556,9576,4113,9972,450,506,891,13037,3340,5242,803,10406,4129,2064,6156,489,2161,515,5331,850,6062,537,1060,2988,12725,3378,803,4445,8637,3097,3105,3505,667,1219,3280,3407,14982,3475,5622,2426,667,16778,3271,608,14803,6316,2591,10662,459,2958,9256,5124,3946,5105,721,864,7797,2037,9569,1370,3530,4872,7143,2761,6188,728,576,521,4762,3713,10164,11390,2148,2680,1238,7978,3856,394,7145,473,2300,5655,518,1063,4423,6442,633,418,5056,5167,841,2680,15092,10964,2266,432,12338,3734,5226,6167,3360,2694,3710,17889,737,3959,13027,700,5516,648,16337,558,3545,13052,1163,648,17916,473,2357,1907,2396,5556,5387,3464,3512,447,391,16709,7325,15852,427,17747,6031,2353,2393,14138,4129,15095,8501,17955,8313,2708,6140,571,3925,382,5602,5002,4187,4374,11021,5269,15272,6012,15510,18026,2496,459,18127,4601,1867,12714,6654,10043,11526,552,2715,10184,16626,654,489,5038,432,14027,17084,795,15218,2430,8808,5269,8366,16945,2601,4090,3734,2732,1393,471,432,3888,5941,435,827,4811,2832,3761,698,1749,372,5346,4795,6566,8743,4171,593,608,5233,2655,1709,1997,8400,4530,8214,2218,1438,7580,1435,2676,4241,810,4679,4933,2093,3808,473,5717,1378,5002,9471,9533,4368,15065,515,628,14740,2371,14792,8257,7955,523,9526,2234,2924,2596,2616,15769,4315,3570,7086,785,1024,2270,4858,3345,544,6980,2981,459,4167,765,473,13182,3872,12182,847,5058,3265,521,2652,1051,5953,438,10389,700,4281,18363,5697,5107,2329,5600,690,844,3360,6726,7139,9960,3042,2507,9659,3959,4497,2912,848,596,3751,9408,8731,15511,2095,4668,941,9961,4939,10093,11434,529,1715,6232,3620,3802,457,5975,3407,2623,5818,17294,5455,6727,2913,4602,1132,4507,1966,7333,480,9513,4914,13813,8996,2228,3685,670,7480,11119,14488,457,3878,6509,2863,1947,4766,6573,336,8975,5869,3669,624,1743,5884,15116,9877,495,6963,7841,9609,3101,905,9478,864,3071,4562,8736,2793,743,1143,2692,12787,3123,6969,1071,2092,918,6960,931,561,7070,965,4504,974,1833,1314,8145,6786,3567,1665,1237,1154,6333,645,13161,702,6564,2800,7357,11235,645,5062,612,2587,1721,17334,710,2321,675,4124,579,6968,1008,3998,506,8067,6411,1013,3072,1333,6962,364,7451,4871,4544,705,5241,10395,932,4579,1362,6738,2371,5919,964,593,13178,3674,18172,4503,10428,6975,1115,3178,969,997,517,596,7032,774,6352,7806,1186,7016,463,2106,1001,3669,14386,1237,4435,1289,1791,933,3215,880,895,1213,1985,8973,4974,2387,15188,1008,3671,593,4679,3332,828,574,3385,976,8128,3411,1728,9257,1118,2051,10454,6539,1170,1069,638,1225,1008,5559,3084,1105,9090,7392,2578,1993,691,7306,561,631,15717,596,1116,6168,587,3648,1760,1046,10636,9952,493,1103,9609,673,555,969,3724,1351,654,868,6026,816,6542,1408,772,6479,1154,7487,5245,3906,4357,827,722,4047,1024,18293,4793,925,1745,863,459,8093,666,14542,821,958,802,524,828,7988,505,936,684,741,730,421,13903,3478,698,7910,1262,2310,854,698,7758,6532,1623,3895,3355,2391,1689,1872,2466,3922,9541,648,5070,4038,1080,9574,4381,12008,7600,827,5799,13068,1656,593,932,1607,6108,12455,766,1935,4718,608,850,1813,1082,1833,1686,900,707,8721,12862,4528,10396,8485,3300,5532,665,7222,7805,905,2528,6470,727,582,1750,2898,1899,2074,764,492,16314,1031,7167,4296,4002,487,4200,701,7128,4281,6669,4724,7466,596,1116,2331,1442,2437,545,11192,4543,4478,4269,462,730,5908,3183,15092,702,3923,1286,1193,13406,8895,537,12916,4150,3852,830,707,2259,1092,4653,1050,3427,687,13420,2042,15797,15105,730,716,5743,1586,522,3307,6743,692,1367,1318,798,6982,4737,9243,579,854,1228,3296,1797,1103,14433,1206,902,802,545,523,10389,8752,765,945,988,1642,1935,434,2414,1875,10082,765,3555,2376,816,6105,6108,2338,3176,2352,2042,2484,898,10038,5958,16297,713,6407,557,844,3037,5610,579,3774,7695,1254,2936,5421,836,1121,1289,7372,1055,1273,752,795,17987,1046,2298,6112,561,734,1081,18607,1187,605,988,739,2137,438,1172,3266,7287,957,3431,861,9734,4062,2856,14593,4815,1656,544,618,4630,433,14201,936,5281,730,2129,645,921,963,7603,1746,1583,7611,802,574,1565,7079,577,764,1244,880,4106,1263,788,561,1641,422,2208,2950,814,1595,2161,752,9556,1569,1607,1138,3015,2801,776,1056,11530,3535,523,1340,9905,7081,16492,6313,11255,1061,2139,891,6296,1737,3502,730,675,7010,12430,7443,833,990,789,462,1151,737,5863,3700,391,616,743,540,819,765,1810,5167,5996,548,3114,743,6486,2051,2479,1121,789,10228,13034,7507,743,1800,6239,6663,10428,4879,9537,3121,943,672,5497,1061,18257,12821,6648,666,878,821,895,3124,741,5919,6397,1939,1343,2290,1949,1091,6438,553,645,2005,1327,2708,684,2647,13720,5002,3437,7199,5242,12831,2206,855,2130,1433,8922,1114,4653,4498,1819,646,666,561,942,504,4309,3752,2616,2960,7155,1363,1229,7026,2486,3104,17513,2443,4471,2810,1259,545,9025,8233,3606,12081,854,10091,1235,761,898,2482,1599,787,561,982,13250,982,1883,3377,854,7049,1008,4484,878,2428,4877,1027,2041,422,4045,540,4242,2338,2357,765,2512,917,1740,1556,1136,1264,4584,7220,1103,2095,645,408,863,17650,596,1408,9094,828,7667,3626,993,2482,684,838,1878,1172,4900,4718,1781,2776,5581,7290,802,2893,990,936,2035,6793,4666,1147,2442,600,6743,531,4084,5439,1125,2442,1607,3966,6239,7414,4210,1166,10646,8501,1052,878,16733,847,11096,802,8032,1087,1949,5044,612,772,7059,16336,1026,12437,1229,10688,4195,746,776,6989,435,1257,946,844,1659,5698,2489,4538,11577,3172,1009,4355,5256,751,2674,719,5522,7577,15144,956,684,1881,698,1020,2550,5701,4297,802,1235,492,1916,855,1909,1893,3549,15303,462,7548,2972,1076,13540,507,6499,16854,14745,3667,915,787,7583,2496,6019,2473,4672,1914,2386,878,6169,8057,3738,891,1179,5653,13978,1046,7848,9710,650,895,2347,9741,820,462,796,877,5967,726,1792,2451,6377,854,408,708,11544,6104,3035,9691,5600,1114,2877,797,6539,8316,2599,8794,15568,1949,1266,432,17724,868,544,4973,776,921,973,1875,6928,984,579,491,1914,13940,579,1727,1911,2234,1847,407,14024,2064,1624,3012,9137,783,1689,5484,2382,5739,8113,9158,6115,1052,7247,2115,707,855,5408,7942,645,2467,14674,11376,5792,10947,2215,2819,921,1624,1974,645,1935,702,13994,5898,645,1193,1381,1622,7076,877,1367,873,1721,612,8121,816,12947,1624,3656,5044,4277,6295,1114,3589,847,438,813,1173,945,8596,638,957,15843,6499,3614,2233,1061,7467,2131,9214,649,18741,2016,4473,15197,15562,2012,1148,1689,5967,2040,3478,789,18419,477,979,706,1698,2045,984,2018,3398,827,4565,8349,1059,4627,743,8213,8184,10302,984,863,4771,720,3402,15466,13912,8280,10388,1637,4276,7875,12958,477,1648,1715,1198,3323,624,2815,2285,833,4318,1035,15977,6733,926,1694,1289,918,7258,6377,4887,1076,6160,3089,10053,695,11584,13787,5940,684,2753,9013,4416,503,394,2104,5006,1881,9525,5088,6777,2295,711,687,743,2150,1056,7655,2095,548,434,1098,678,5183,16599,4268,882,3985,4096,1046,1689,1662,652,3506,1401,816,2352,9541,6857,596,7466,1624,5759,1838,666,540,9230,949,526,2271,1824,421,936,8151,960,1644,7375,15134,5480,2344,1935,5169,784,2919,456,3350,4158,2646,2314,1066,906,2153,5759,8921,2481,945,984,817,1384,1083,8932,432,1655,828,526,984,7173,593,2724,6500,2657,8281,1421,17579,1179,1949,833,2551,4127,13293,505,449,3673,673,2686,7931,5025,6618,863,10226,394,1392,505,4284,868,689,9289,4847,432,828,10886,2375,8639,5294,5946,2009,965,2834,1443,906,7589,707,12848,758,1400,622,7351,952,1842,990,2949,4466,4064,1018,571,530,970,1007,1956,8868,11765,12185,447,993,710,1374,803,1662,1359,670,5443,3205,10706,2822,17153,13098,3250,2452,720,4123,4196,4044,16558,734,1848,1821,1015,4466,733,507,1807,3664,4214,942,950,3345,12707,713,2719,1775,720,2551,805,5364,734,663,1200,7662,436,14561,1777,1752,10308,1786,5817,2699,851,2764,1151,3634,6125,2583,487,805,732,4189,1892,3965,3298,1323,4946,2883,2647,499,4956,485,1140,17216,734,668,3284,3304,1919,1814,548,3312,683,732,911,12648,720,1171,4435,1383,1190,2508,780,3871,2579,574,669,5265,10800,942,4712,1848,777,2518,1002,1015,3965,3236,793,5880,759,756,756,9009,741,706,5198,2224,2863,9173,781,1668,7453,6056,815,933,2670,4751,1196,1399,3045,5526,552,3468,4764,1963,669,3002,1743,1662,3231,550,3465,5804,1050,3593,675,3454,766,653,3035,2841,2841,5805,4639,2227,7114,1235,16137,2426,2847,8716,10562,5841,666,3968,1945,2426,2681,530,1033,10580,1956,847,1882,13631,4068,1067,1175,3057,1063,1167,5793,4633,6563,2239,2562,2865,2567,4044,2078,4949,2804,1893,1232,3296,717,1610,933,791,952,949,4697,756,2827,4946,947,552,5775,6443,6066,7305,2200,17659,1064,3562,2610,7812,5443,593,2821,4284,663,1166,4971,5980,972,574,2211,2247,515,563,552,530,12338,1387,2863,972,1172,1095,5042,791,716,4538,3347,2546,5533,1687,1268,4612,636,1993,948,687,925,2407,805,2475,681,972,2415,3448,5164,1752,734,4588,1111,8930,2328,3462,3791,8138,2604,4564,926,684,5872,3304,777,3006,4583,8312,1034,6429,805,2726,644,1333,1078,1326,3871,9836,3799,734,7001,1190,1928,5293,1146,1257,1664,610,1600,816,4514,777,2319,701,4096,1002,6196,2037,15992,3253,9738,2348,2564,3763,3353,5671,1419,709,814,10607,610,2826,7677,3160,589,876,1644,758,10907,3049,571,3214,2880,2331,681,1845,7487,3618,738,552,799,12738,1569,1031,2686,3173,2475,3195,2313,804,2387,5939,670,1074,3135,571,3078,4223,3685,507,4295,4682,681,990,11078,593,612,3740,1356,5005,7679,3473,5443,2933,554,6020,801,890,642,608,2878,2926,1002,4191,972,650,492,4032,8341,827,756,6776,663,1690,12981,766,651,4447,10115,1092,1963,1323,362,4068,734,758,1662,4916,3278,462,697,622,1202,13111,2718,810,1431,1030,756,1822,492,808,1610,1074,10512,2467,758,734,622,4312,4725,8119,758,14236,13588,763,1433,16872,1882,1353,3489,1619,404,1551,15334,3937,1799,633,626,1923,1190,756,1822,713,732,2939,1814,942,6075,4811,8818,593,1272,794,5422,3107,532,803,7966,5639,669,447,1687,3135,4633,972,1015,1752,1963,2642,2217,4427,593,7127,1806,1845,589,4253,2360,774,12799,3077,1857,1569,1786,10090,875,1720,12581,790,1882,941,683,2033,880,1177,4229,1771,2563,3284,548,911,1698,2866,1207,653,10051,13603,5897,855,9720,1002,1035,853,3618,6857,4365,4168,413,608,868,1015,8020,3075,2484,414,11147,5755,852,5821,1133,462,781,1030,791,2015,942,710,492,478,454,1612,2811,5979,1063,911,967,4476,1873,1410,3839,5134,4869,7257,373,1132,652,710,6319,710,13162,893,1924,17590,2288,1628,571,1401,4693,1257,1264,3382,1991,669,794,4517,733,4732,11512,2752,588,17761,5219,686,11825,720,538,1240,1899,2797,2578,13786,3802,6250,1115,4670,12338,4973,1381,1981,612,1757,7333,6426,2066,898,7440,1184,11146,4584,12146,3011,992,905,540,2330,607,1867,4060,5468,8818,4689,3624,4212,4890,4821,1552,1194,10345,775,5893,4819,449,1155,8069,907,9081,534,2505,4129,485,5334,668,8829,2157,17073,7438,776,5125,700,7513,896,17882,3024,9642,4740,5845,2898,1370,6453,5610,1057,1024,11088,14968,700,6013,726,5013,5154,13445,5018,4101,523,954,17849,11322,1017,12196,5544,4578,5031,4924,1208,9697,11177,6019,3437,1838,4816,4749,6405,521,2313,828,1011,1332,8260,1205,5363,526,9780,505,15324,8654,806,505,7246,18648,1268,1741,18118,2827,3220,12121,4873,491,5882,6036,2846,1671,545,1395,2368,648,12829,2375,10546,1655,2912,4306,4435,407,12677,537,645,4746,6871,921,5102,421,2172,579,708,1010,775,590,628,5550,5324,2138,2489,16737,15974,4038,4149,720,6288,18795,2269,1169,2303,15035,1020,2420,842,5171,4559,5063,776,8261,2935,7849,18108,3447,6459,4309,3987,6340,4513,6199,6406,8275,18440,4037,2327,14543,4687,8529,16111,995,1749,5523,524,5219,12117,576,4081,3888,596,584,12907,3394,433,5190,967,3862,625,2870,3472,761,5492,2517,441,4109,5718,725,645,12082,1944,3190,6793,2106,1439,6547,802,4514,6296,4704,3089,10134,1427,18398,4007,2184,5291,8541,526,707,689,2113,665,3472,5100,2394,628,4381,4113,7534,1599,683,4129,912,2367,13387,631,1365,878,5824,4973,625,362,4566,12459,614,1628,1292,647,9694,2596,6042,4861,8624,854,7617,8815,5242,608,788,600,729,1096,1299,1820,7929,1817,4569,2992,1584,5750,730,941,16353,5433,4498,2590,7673,1140,3979,3884,2760,749,16582,17634,7999,12121,464,713,646,871,526,14603,3180,482,7747,2956,2801,3819,1154,6798,3603,4535,877,3167,4001,4872,625,5219,7415,1863,3167,686,2699,573,507,13629,2517,13068,3924,675,8014,623,3160,1270,514,579,4033,965,2751,1811,517,3996,4788,707,4309,16390,1771,4186,7125,4341,2207,3352,12308,8709,12747,1268,2636,9452,12874,4243,2500,4191,5267,4980,789,2819,6588,5143,4596,12171,3557,4639,5818,4724,2293,2444,3890,4323,1111,10546,540,9896,5565,1177,1115,6814,551,17235,9572,3324,5312,2594,706,10274,4233,867,6089,789,473,10886,568,648,2644,936,6797,853,1445,645,1808,10023,698,5236,8286,633,2710,828,2648,5468,5561,5593,2176,596,894,4389,1799,15618,5103,6770,9253,4200,628,10584,1613,17247,5125,1094,3274,421,484,2534,4619,5423,3967,5868,5220,6337,3989,2536,489,5706,4749,4830,6781,2583,727,1591,1412,4937,11796,15467,4140,429,4702,5838,4578,496,5698,921,6037,4514,5622,763,6250,7567,473,965,878,7273,1128,1436,3095,4959,4039,12485,4883,4320,4155,1694,778,3832,683,4826,4418,2949,2549,12720,4432,5296,1956,8774,1808,2233,6814,6838,393,521,1080,1686,6132,2368,700,6140,4965,5437,4559,1187,7774,4389,16677,10739,2781,2815,1045,7577,608,868,539,2145,505,5852,907,5218,521,3775,5204,5938,4252,5050,702,668,4475,9523,13099,579,2209,2066,4558,4826,943,3348,1024,614,4830,5140,900,648,526,6578,2945,11105,2923,4290,432,2020,18426,912,7918,776,936,1706,2784,1879,6088,13502,4749,561,2048,6564,1048,5376,1454,743,5010,919,9768,2425,4657,1111,2112,1983,5412,11360,2428,568,628,5312,878,625,12071,1827,16690,647,478,5171,10424,833,938,862,11200,794,1329,3854,1821,537,1577,5932,485,868,15857,1433,7098,3296,1235,10012,684,4688,4088,827,729,605,3249,6619,5799,4484,1024,7277,827,1187,602,6867,8686,1450,5688,5950,5189,6480,7476,12809,1289,7633,1715,3640,5171,12047,12923,932,1161,5592,3775,4642,764,7400,17522,8931,14768,2588,689,10337,18440,5670,1616,4520,2096,6588,5914,610,1600,3869,3105,906,17569,1158,1412,838,3030,4772,1573,898,480,2481,1402,6904,3676,6344,1011,5241,16643,3290,4379,435,2939,11975,6588,3485,3781,4412,17849,898,3731,491,3920,4662,1569,2157,12219,4395,4789,941,1846,8951,2222,511,4659,3296,730,10738,620,3160,10738,2548,1957,675,2100,5783,3183,4537,4939,3921,3730,15451,7466,2484,786,2178,4827,383,698,936,3387,4688,448,13949,1401,8127,5450,545,689,2828,4578,18565,1239,3142,5247,17175,1200,12418,8569,1124,1554,10351,2329,2575,3180,2329,918,702,1815,4118,755,975,1420,1041,1013,470,811,530,2112,2453,1153,2271,4513,7703,1154,813,9474,2569,6221,3590,930,898,2753,5599,923,1227,401,1374,16409,1668,999,9882,584,3311,5372,530,2871,622,3307,16779,7665,864,3948,2008,2239,3026,2307,687,6852,614,766,505,6517,3150,3884,4092,5007,789,2467,4480,1153,3176,1682,12968,5330,2040,6666,5359,1304,951,984,1053,477,2511,2838,3295,1238,1053,4570,2854,3942,4232,7274,5174,2026,5065,1910,1024,1758,829,3107,3585,1088,1244,675,5132,14179,596,3050,6518,2280,3063,900,1041,2010,470,842,2002,4392,2345,1060,434,1250,984,1781,7006,6849,1042,846,734,4504,1554,1712,745,13254,1744,4256,2753,9281,1564,4429,768,673,6091,3133,2901,511,1372,8303,7182,3175,757,4257,2817,10314,3595,1153,478,680,7676,1062,1624,3024,1041,10781,1098,3380,2879,4398,2973,4623,3723,1274,1819,1359,12265,782,540,1153,658,702,1407,1717,872,765,2347,2030,540,14931,1167,1176,956,949,999,675,5268,620,1268,1334,740,658,10285,2629,3914,641,765,4439,596,6989,2442,4797,808,838,1013,6245,1637,1656,684,1343,1008,1223,10113,1014,1079,3941,1951,15662,6103,3360,1056,9205,2556,4203,1624,1758,3478,1815,4807,3192,608,5351,12210,773,1003,5301,1975,1046,1207,658,596,507,3278,2570,12754,9796,928,2590,5330,4068,1097,723,2369,678,2906,851,619,2143,4751,3800,755,811,743,1767,2949,814,949,755,1637,1348,1435,7131,1984,3801,872,4525,1708,2673,11871,680,526,945,775,984,2529,12000,598,789,419,3506,1013,767,10388,2501,4665,2711,2447,872,2537,4077,4031,7190,7408,13320,693,2656,3026,5250,1614,4183,4766,900,9140,1982,564,3965,1207,5393,1813,781,1059,3381,4760,3063,4562,1607,657,731,834,1075,8073,639,1365,1662,6892,3819,6751,4440,1897,942,4620,8982,13015,805,15649,5319,2041,5002,3249,696,1343,984,642,1559,417,660,3660,3304,698,7145,722,1281,1003,1738,7533,1316,1206,6425,2999,1624,3758,683,969,8698,1004,3601,620,4875,12499,1438,1580,5226,4332,2889,814,2753,4077,16650,1038,1632,2891,3455,2282,11688,576,1128,734,521,1000,449,10283,3293,1153,707,658,738,10935,17825,3462,2753,872,2753,5163,6335,4309,470,987,3250,745,945,11214,1053,3429,900,1000,4506,951,3163,2399,1079,11301,1758,9157,4315,3177,737,1073,1240,865,14482,1621,2576,3061,852,1035,596,5599,4788,651,5645,1334,958,1921,1030,633,465,1282,656,2112,569,6534,1668,1030,4844,766,745,4229,670,5430,811,17871,2711,988,767,844,816,1307,675,4353,4742,4311,890,620,1000,579,969,2125,1580,1875,1752,956,3256,2897,526,4321,6737,3370,612,9680,4286,10313,1211,3864,3342,11586,11279,1662,953,4751,1443,612,5973,8408,435,789,1438,576,984,1343,3006,12236,963,3291,1816,5012,2585,657,6132,1003,5127,1389,1890,8220,3136,12467,1649,4265,3752,1273,999,837,3105,635,6806,842,2453,1785,743,1018,1001,9705,1903,912,1637,10851,789,1410,2735,4939,2913,1801,1577,2306,1043,4379,2647,864,3464,1018,7560,4026,975,1004,463,6682,975,1443,8836,723,9513,2441,552,526,984,1656,1809,1214,4183,9089,2788,12644,5058,532,3895,805,705,544,5229,702,1829,1875,3924,872,4738,419,1170,1662,3136,767,646,702,10915,6563,1613,713,2324,2940,6516,17688,2442,801,900,2516,16277,3941,2688,2498,7251,8888,4472,844,2923,1827,1154,1817,4688,470,1841,1894,1809,1002,658,384,875,2618,1250,4853,631,1125,1665,723,906,1303,1170,1274,3484,7963,900,6335,3184,984,4791,4620,1160,1367,734,4580,1918,745,773,702,1410,7094,872,714,544,18067,1607,2657,427,518,6285,1332,9904,8972,3744,646,1645,1238,1743,2511,873,3519,2617,1717,743,1625,844,1355,1949,1656,10993,1644,4164,705,16169,3226,984,1046,4478,8492,1116,779,1085,1815,10539,3748,1700,734,743,852,755,3435,1238,743,2573,702,18176,552,949,837,11382,1079,2564,7747,3553,3247,2789,1888,521,2301,11554,1082,675,743,477,1048,3570,842,1778,3197,1656,18236,15984,526,3833,1154,499,944,6473,917,16957,1419,3917,3192,575,2079,1090,3012,1243,1637,3889,3340,844,4562,3595,4196,8674,935,944,658,1316,477,13661,658,1760,4921,596,2802,765,603,1694,945,1415,1395,7589,1754,1187,2398,1865,1316,9278,5042,855,1745,5067,4864,680,544,862,6403,7260,992,1248,5458,421,758,1245,9058,1243,548,3795,14386,755,3091,3697,4576,844,779,1238,1727,8167,4563,11644,6335,1624,625,1279,2765,1012,2135,942,400,3103,1184,972,4936,14527,8994,1254,800,872,4482,2427,921,576,1402,3189,2857,716,3293,603,7610,1424,485,4030,1737,674,692,810,3053,2229,596,2814,10888,1884,2826,737,5381,844,789,16261,10211,14092,876,17694,607,6416,14465,1436,760,6887,703,4844,1882,16240,1803,2839,1193,5292,694,857,1594,662,1172,4108,1107,2239,491,868,2645,682,664,9203,3011,2643,4981,2291,2656,15247,2331,1348,6278,828,5410,10804,664,6301,1656,13853,4774,642,10197,2799,453,825,5889,4282,12343,1103,1744,2039,3376,15968,5938,3950,1749,2777,1723,802,2686,5420,1698,583,4594,6701,4025,1389,14247,12071,6075,9043,4844,5046,1116,460,15688,840,3005,905,9092,2198,828,1643,5152,9408,1050,644,879,2167,2036,2717,2445,4403,2745,13325,711,16670,17869,2386,2381,770,1103,6652,675,8688,879,939,2808,4743,2043,470,7372,587,1853,9797,6467,1577,2196,1800,2976,2102,697,7741,5215,1766,764,2753,7632,686,2607,17203,7781,6458,1552,10436,2431,12320,4162,854,9122,14833,1870,886,487,2932,2839,1059,2100,802,678,12734,1953,3920,11654,5378,3759,2074,2874,731,1702,4508,1441,18578,2874,1323,1142,1115,3090,1605,2212,3264,12621,5174,4926,1115,4140,4095,599,666,840,3663,4755,15773,886,761,6271,1591,9596,1865,3502,3518,971,4328,5729,802,2264,2369,6288,919,2088,904,2017,1748,3789,1089,3039,673,2624,5606,2079,1040,773,2553,2795,1273,645,2527,487,4972,17279,620,17455,3082,5151,914,14483,684,1666,776,530,2762,1291,7056,957,2301,8879,12175,645,4427,1356,6047,2516,1046,1436,5937,5894,16256,10937,4596,478,766,738,2889,1901,652,15841,852,6391,731,518,11666,3816,9218,6097,1174,1805,1187,2177,685,13853,1627,3613,393,11051,5505,867,2406,1588,4497,10494,904,576,13215,8627,2314,17366,4082,524,3096,2290,3991,504,1837,679,8334,1192,3321,2960,7893,3368,597,2298,1968,3018,625,1356,8430,7752,975,746,2372,2642,1914,7123,10962,1026,5021,679,583,906,1240,863,421,4832,3043,402,4851,783,6073,1358,2117,456,3787,2801,781,1569,1117,906,6097,906,1691,4446,4969,7179,3010,3453,3774,4181,645,7056,1682,2826,1053,475,15851,1026,1961,1760,5292,886,9419,4298,10742,968,2801,2025,731,1050,2404,1442,4081,530,4131,2219,2410,2839,421,4221,5087,9533,1031,2790,5164,802,790,3282,18395,1436,2261,1279,7486,998,828,2862,1059,2392,2328,492,2807,1142,1792,4586,2100,5817,2230,3750,548,2400,1031,1741,3672,724,2269,15955,12141,4063,14386,1031,4690,5557,600,4899,3499,746,1676,814,1085,13872,5919,5505,6676,780,814,6456,13653,871,2564,6244,4363,593,472,776,6455,1662,921,1354,2052,14066,2699,624,11549,3811,754,1168,2016,490,897,3787,1657,957,2264,2777,906,4480,3062,7870,4004,3198,8114,2320,3734,394,4414,4851,5010,842,828,2400,2838,597,9581,1115,9641,1960,4282,2085,2443,4162,945,4729,4238,10217,573,1562,1095,956,796,4989,5819,2309,616,789,6662,3436,506,3653,3468,1271,1607,453,8816,1107,2312,706,12677,2275,886,645,9461,2258,7677,737,505,2795,5141,487,1007,1564,4279,17534,4228,2130,776,5816,5288,645,2016,1236,540,1016,7706,18429,2287,4006,1812,6925,945,447,931,926,14217,2354,642,1026,3806,7238,2011,8476,761,1436,518,613,6204,530,1373,6662,5142,5889,1915,802,2184,6762,3996,7275,558,2137,8505,988,3796,625,668,4953,949,1196,1974,6825,2642,705,9552,10577,3405,645,11233,2469,931,2686,827,828,3655,7350,16116,4124,4230,8983,5308,6108,801,1897,4486,2359,2936,8964,4554,557,802,6094,2319,669,1132,11946,1571,830,914,612,1989,773,800,470,12549,918,3336,6899,1744,645,524,2744,1799,14199,2717,748,2190,679,827,1333,1129,1562,4719,928,394,1356,1026,2326,7258,1351,1428,3913,554,7014,819,2805,1676,842,1247,2227,3437,5825,1600,5925,14196,4137,1939,11525,2348,2200,14294,1021,2751,2381,2877,5174,6751,11957,5119,5557,583,787,7343,516,10204,652,5017,886,1745,3701,4412,1221,2805,7003,4989,3763,5646,4508,684,608,863,3363,2359,2082,663,790,6261,14611,1139,889,2394,18055,4383,3590,3162,706,6686,13853,3002,7162,954,5638,815,682,3544,4480,1814,863,4536,1871,4939,1085,13080,1308,603,633,737,3064,1621,2158,828,461,3810,10032,4497,419,2373,2382,945,1142,3013,6039,8314,7334,5241,4853,419,481,2316,3118,6885,11382,839,17425,4899,4013,2423,4308,2446,12610,3350,6195,945,1591,5038,4319,1550,3226,4816,625,1358,1569,5487,17029,4468,517,2237,2058,10771,3704,990,1654,394,1615,3694,1857,873,2977,583,983,2469,5288,3745,3749,1778,9377,4770,612,5786,2345,754,4180,1561,4712,3562,641,9245,2658,8820,666,2400,1777,9281,13646,548,793,3546,1076,1680,538,787,1240,1322,1007,5292,2356,1881,2494,1065,926,1830,1122,15031,6330,673,13248,1215,1192,8216,1410,2401,18159,1122,994,688,1394,470,870,499,1131,3599,3455,3061,3801,13043,384,3601,1666,1940,12862,494,2854,533,12196,1836,4516,2148,3080,6079,3399,4194,7136,8662,7162,1626,648,1224,4200,4824,965,3181,7942,1832,7368,784,552,13564,368,4611,12832,4754,5518,566,11764,9271,978,3528,10309,6702,9882,9380,7589,6966,4315,8183,8138,4816,6982,506,421,10165,9728,3312,2104,11106,3299,2036,1062,4173,4686,489,994,2705,7207,2295,1814,8013,574,765,7811,3465,7677,4588,418,15246,2162,12554,2442,1179,5455,1953,15714,11576,15109,3755,830,7553,7844,5761,8848,12267,8540,7637,4292,2258,5016,9704,8227,875,591,7553,15110,2047,2150,6851,5707,9510,4838,3603,8110,5893,3669,1821,4838,1606,3445,1125,4222,10401,469,915,851,16538,5606,1986,5569,5067,790,5504,488,5624,16062,335,7550,5412,15231,4742,4854,4032,2873,2257,13499,1958,1165,856,3496,8048,4508,7899,459,1588,1869,17920,12381,5430,393,17209,1415,828,708,6750,7903,1864,4619,983,7730,394,830,15824,3210,810,9095,12845,3783,694,11188,13782,4144,9555,5160,5140,1657,581,9793,2184,3266,6201,2278,450,11189,4492,4385,5969,3462,4191,494,411,13543,1902,4173,662,6966,8104,1129,617,727,11854,7515,8107,5252,14527,464,8930,3145,2365,4779,2159,4064,7943,16198,3790,5264,1896,5450,1782,18706,613,405,957,454,3418,1592,523,6996,4582,13092,810,4189,830,11392,7518,7308,5912,6629,17891,1949,3740,7455],"hoverinfo":"y","type":"box","fillcolor":"rgba(255,255,255,1)","marker":{"opacity":null,"outliercolor":"rgba(0,0,0,1)","line":{"width":1.88976377952756,"color":"rgba(0,0,0,1)"},"size":5.66929133858268},"line":{"color":"rgba(51,51,51,1)","width":1.88976377952756},"showlegend":false,"xaxis":"x","yaxis":"y","frame":null}],"layout":{"margin":{"t":28.1765601217656,"r":7.30593607305936,"b":42.130898021309,"l":54.7945205479452},"plot_bgcolor":"rgba(235,235,235,1)","paper_bgcolor":"rgba(255,255,255,1)","font":{"color":"rgba(0,0,0,1)","family":"","size":14.6118721461187},"xaxis":{"domain":[0,1],"automargin":true,"type":"linear","autorange":false,"range":[0.4,7.6],"tickmode":"array","ticktext":["D","E","F","G","H","I","J"],"tickvals":[1,2,3,4,5,6,7],"categoryorder":"array","categoryarray":["D","E","F","G","H","I","J"],"nticks":null,"ticks":"outside","tickcolor":"rgba(51,51,51,1)","ticklen":3.65296803652968,"tickwidth":0.66417600664176,"showticklabels":true,"tickfont":{"color":"rgba(77,77,77,1)","family":"","size":11.689497716895},"tickangle":-0,"showline":false,"linecolor":null,"linewidth":0,"showgrid":true,"gridcolor":"rgba(255,255,255,1)","gridwidth":0.66417600664176,"zeroline":false,"anchor":"y","title":{"text":"color","font":{"color":"rgba(0,0,0,1)","family":"","size":14.6118721461187}},"hoverformat":".2f"},"yaxis":{"domain":[0,1],"automargin":true,"type":"linear","autorange":false,"range":[-588,19718],"tickmode":"array","ticktext":["0","5000","10000","15000"],"tickvals":[0,5000,10000,15000],"categoryorder":"array","categoryarray":["0","5000","10000","15000"],"nticks":null,"ticks":"outside","tickcolor":"rgba(51,51,51,1)","ticklen":3.65296803652968,"tickwidth":0.66417600664176,"showticklabels":true,"tickfont":{"color":"rgba(77,77,77,1)","family":"","size":11.689497716895},"tickangle":-0,"showline":false,"linecolor":null,"linewidth":0,"showgrid":true,"gridcolor":"rgba(255,255,255,1)","gridwidth":0.66417600664176,"zeroline":false,"anchor":"x","title":{"text":"price","font":{"color":"rgba(0,0,0,1)","family":"","size":14.6118721461187}},"hoverformat":".2f"},"shapes":[{"type":"rect","fillcolor":null,"line":{"color":null,"width":0,"linetype":[]},"yref":"paper","xref":"paper","x0":0,"x1":1,"y0":0,"y1":1}],"showlegend":false,"legend":{"bgcolor":"rgba(255,255,255,1)","bordercolor":"transparent","borderwidth":1.88976377952756,"font":{"color":"rgba(0,0,0,1)","family":"","size":11.689497716895}},"hovermode":"closest","barmode":"relative"},"config":{"doubleClick":"reset","modeBarButtonsToAdd":["hoverclosest","hovercompare"],"showSendToCloud":false},"source":"A","attrs":{"13c6a2f07ad9c":{"x":{},"y":{},"type":"box"}},"cur_data":"13c6a2f07ad9c","visdat":{"13c6a2f07ad9c":["function (y) ","x"]},"highlight":{"on":"plotly_click","persistent":false,"dynamic":false,"selectize":false,"opacityDim":0.2,"selected":{"opacity":1},"debounce":0},"shinyEvents":["plotly_hover","plotly_click","plotly_selected","plotly_relayout","plotly_brushed","plotly_brushing","plotly_clickannotation","plotly_doubleclick","plotly_deselect","plotly_afterplot","plotly_sunburstclick"],"base_url":"https://plot.ly"},"evals":[],"jsHooks":[]}</script> --- # **ggiraph** with **ggplot2** style A new library that allows to keep the aesthetics than origin **ggplot2** ```r library(ggiraph) data <- mtcars data$carname <- row.names(data) gg_point = ggplot(data = data) + geom_point_interactive(aes(x = wt, y = qsec, color = disp, tooltip = carname, data_id = carname)) + theme_minimal() girafe(ggobj = gg_point, options = list(opts_sizing(rescale = TRUE, width = .3))) ``` <div class="girafe html-widget html-fill-item-overflow-hidden html-fill-item" id="htmlwidget-931d09d58501d3eb0318" style="width:504px;height:504px;"></div> <script type="application/json" data-for="htmlwidget-931d09d58501d3eb0318">{"x":{"html":"<?xml version=\"1.0\" encoding=\"UTF-8\"?>\n<svg xmlns='http://www.w3.org/2000/svg' xmlns:xlink='http://www.w3.org/1999/xlink' class='ggiraph-svg' role='img' id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf' viewBox='0 0 504 504'>\n <defs id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_defs'>\n <clipPath id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_c1'>\n <rect x='0' y='0' width='504' height='504'/>\n <\/clipPath>\n <clipPath id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_c2'>\n <rect x='37.79' y='5.98' width='394.77' height='463.44'/>\n <\/clipPath>\n <\/defs>\n <g id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_rootg' class='ggiraph-svg-rootg'>\n <g clip-path='url(#svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_c1)'>\n <rect x='0' y='0' width='504' height='504' fill='#FFFFFF' fill-opacity='1' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.75' stroke-linejoin='round' stroke-linecap='round' class='ggiraph-svg-bg'/>\n <\/g>\n <g clip-path='url(#svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_c2)'>\n <polyline points='37.79,423.28 432.56,423.28' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='0.58' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='37.79,322.97 432.56,322.97' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='0.58' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='37.79,222.65 432.56,222.65' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='0.58' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='37.79,122.34 432.56,122.34' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='0.58' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='37.79,22.03 432.56,22.03' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='0.58' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='54.54,469.42 54.54,5.98' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='0.58' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='146.30,469.42 146.30,5.98' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='0.58' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='238.06,469.42 238.06,5.98' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='0.58' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='329.83,469.42 329.83,5.98' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='0.58' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='421.59,469.42 421.59,5.98' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='0.58' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='37.79,373.12 432.56,373.12' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='37.79,272.81 432.56,272.81' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='37.79,172.50 432.56,172.50' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='37.79,72.18 432.56,72.18' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='100.42,469.42 100.42,5.98' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='192.18,469.42 192.18,5.98' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='283.95,469.42 283.95,5.98' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <polyline points='375.71,469.42 375.71,5.98' fill='none' stroke='#EBEBEB' stroke-opacity='1' stroke-width='1.16' stroke-linejoin='round' stroke-linecap='butt'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e1' cx='157.31' cy='350.05' r='1.47pt' fill='#214667' fill-opacity='1' stroke='#214667' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Mazda RX4' data-id='Mazda RX4'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e2' cx='180.71' cy='321.96' r='1.47pt' fill='#214667' fill-opacity='1' stroke='#214667' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Mazda RX4 Wag' data-id='Mazda RX4 Wag'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e3' cx='129.79' cy='242.21' r='1.47pt' fill='#193652' fill-opacity='1' stroke='#193652' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Datsun 710' data-id='Datsun 710'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e4' cx='211.91' cy='200.58' r='1.47pt' fill='#316692' fill-opacity='1' stroke='#316692' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Hornet 4 Drive' data-id='Hornet 4 Drive'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e5' cx='232.56' cy='321.96' r='1.47pt' fill='#4289C1' fill-opacity='1' stroke='#4289C1' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Hornet Sportabout' data-id='Hornet Sportabout'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e6' cx='234.39' cy='161.46' r='1.47pt' fill='#2B5B83' fill-opacity='1' stroke='#2B5B83' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Valiant' data-id='Valiant'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e7' cx='244.49' cy='381.15' r='1.47pt' fill='#4289C1' fill-opacity='1' stroke='#4289C1' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Duster 360' data-id='Duster 360'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e8' cx='209.62' cy='172.5' r='1.47pt' fill='#1F4262' fill-opacity='1' stroke='#1F4262' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Merc 240D' data-id='Merc 240D'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e9' cx='205.95' cy='27.04' r='1.47pt' fill='#1E405F' fill-opacity='1' stroke='#1E405F' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Merc 230' data-id='Merc 230'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e10' cx='232.56' cy='257.76' r='1.47pt' fill='#22486A' fill-opacity='1' stroke='#22486A' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Merc 280' data-id='Merc 280'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e11' cx='232.56' cy='227.67' r='1.47pt' fill='#22486A' fill-opacity='1' stroke='#22486A' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Merc 280C' data-id='Merc 280C'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e12' cx='290.37' cy='302.9' r='1.47pt' fill='#346C9A' fill-opacity='1' stroke='#346C9A' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Merc 450SE' data-id='Merc 450SE'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e13' cx='259.17' cy='292.87' r='1.47pt' fill='#346C9A' fill-opacity='1' stroke='#346C9A' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Merc 450SL' data-id='Merc 450SL'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e14' cx='263.76' cy='272.81' r='1.47pt' fill='#346C9A' fill-opacity='1' stroke='#346C9A' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Merc 450SLC' data-id='Merc 450SLC'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e15' cx='398.65' cy='273.81' r='1.47pt' fill='#56B1F7' fill-opacity='1' stroke='#56B1F7' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Cadillac Fleetwood' data-id='Cadillac Fleetwood'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e16' cx='414.62' cy='281.84' r='1.47pt' fill='#54ADF1' fill-opacity='1' stroke='#54ADF1' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Lincoln Continental' data-id='Lincoln Continental'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e17' cx='407.37' cy='301.9' r='1.47pt' fill='#50A5E7' fill-opacity='1' stroke='#50A5E7' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Chrysler Imperial' data-id='Chrysler Imperial'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e18' cx='118.77' cy='199.08' r='1.47pt' fill='#142D46' fill-opacity='1' stroke='#142D46' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Fiat 128' data-id='Fiat 128'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e19' cx='65.09' cy='246.73' r='1.47pt' fill='#142C45' fill-opacity='1' stroke='#142C45' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Honda Civic' data-id='Honda Civic'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e20' cx='85.28' cy='177.51' r='1.47pt' fill='#132B43' fill-opacity='1' stroke='#132B43' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Toyota Corolla' data-id='Toyota Corolla'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e21' cx='143.09' cy='171.99' r='1.47pt' fill='#1A3A57' fill-opacity='1' stroke='#1A3A57' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Toyota Corona' data-id='Toyota Corona'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e22' cx='239.9' cy='329.49' r='1.47pt' fill='#3B7AAD' fill-opacity='1' stroke='#3B7AAD' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Dodge Challenger' data-id='Dodge Challenger'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e23' cx='232.1' cy='307.92' r='1.47pt' fill='#3875A7' fill-opacity='1' stroke='#3875A7' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='AMC Javelin' data-id='AMC Javelin'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e24' cx='269.26' cy='402.71' r='1.47pt' fill='#4085BC' fill-opacity='1' stroke='#4085BC' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Camaro Z28' data-id='Camaro Z28'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e25' cx='269.72' cy='320.46' r='1.47pt' fill='#4997D4' fill-opacity='1' stroke='#4997D4' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Pontiac Firebird' data-id='Pontiac Firebird'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e26' cx='94.46' cy='227.67' r='1.47pt' fill='#142D46' fill-opacity='1' stroke='#142D46' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Fiat X1-9' data-id='Fiat X1-9'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e27' cx='113.27' cy='338.01' r='1.47pt' fill='#1A3A57' fill-opacity='1' stroke='#1A3A57' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Porsche 914-2' data-id='Porsche 914-2'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e28' cx='55.73' cy='327.98' r='1.47pt' fill='#17324D' fill-opacity='1' stroke='#17324D' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Lotus Europa' data-id='Lotus Europa'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e29' cx='207.78' cy='448.36' r='1.47pt' fill='#4085BD' fill-opacity='1' stroke='#4085BD' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Ford Pantera L' data-id='Ford Pantera L'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e30' cx='171.08' cy='398.2' r='1.47pt' fill='#1E4161' fill-opacity='1' stroke='#1E4161' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Ferrari Dino' data-id='Ferrari Dino'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e31' cx='244.49' cy='443.34' r='1.47pt' fill='#3874A5' fill-opacity='1' stroke='#3874A5' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Maserati Bora' data-id='Maserati Bora'/>\n <circle id='svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_e32' cx='172' cy='242.72' r='1.47pt' fill='#1B3A57' fill-opacity='1' stroke='#1B3A57' stroke-opacity='1' stroke-width='0.71' stroke-linejoin='round' stroke-linecap='round' title='Volvo 142E' data-id='Volvo 142E'/>\n <\/g>\n <g clip-path='url(#svg_0c9c18b4_2e68_4579_b86d_088360a75cbf_c1)'>\n <text x='20.21' y='376.62' font-size='7.2pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>16<\/text>\n <text x='20.21' y='276.3' font-size='7.2pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>18<\/text>\n <text x='20.21' y='175.99' font-size='7.2pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>20<\/text>\n <text x='20.21' y='75.68' font-size='7.2pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>22<\/text>\n <text x='97.37' y='481.79' font-size='7.2pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>2<\/text>\n <text x='189.13' y='481.79' font-size='7.2pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>3<\/text>\n <text x='280.9' y='481.79' font-size='7.2pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>4<\/text>\n <text x='372.66' y='481.79' font-size='7.2pt' font-family='DejaVu Sans' fill='#4D4D4D' fill-opacity='1'>5<\/text>\n <text x='227.91' y='495.52' font-size='9pt' font-family='DejaVu Sans'>wt<\/text>\n <text transform='translate(14.73,251.63) rotate(-90.00)' font-size='9pt' font-family='DejaVu Sans'>qsec<\/text>\n <image x='450.49' y='203.11' width='17.28' height='86.4' preserveAspectRatio='none' xlink:href='data:image/png;base64,iVBORw0KGgoAAAANSUhEUgAAAAEAAAEsCAYAAAACUNnVAAAAmElEQVQ4ja2UQRLDMAgDd3hk/tnX5agcknFaCoa0vnkMCEnGsL12GQhDYDpPwoDrpBFlRDWi/9Wu6AHC9FkLYLrzlhD3oJ3mDvRHRYy7IC8A7bCK8gpWKYDzeQVArDcDyD2YAnSofZvzaOA6Q7Oahp/dmRuVTQ0xU5TGP2o8Waool1obVuv1q8qP9xPvg633XlGLDtfIhNUBcBeA5ss0BXMAAAAASUVORK5CYII=' xmlns:xlink='http://www.w3.org/1999/xlink'/>\n <text x='473.75' y='286.66' font-size='7.2pt' font-family='DejaVu Sans'>100<\/text>\n <text x='473.75' y='265.18' font-size='7.2pt' font-family='DejaVu Sans'>200<\/text>\n <text x='473.75' y='243.7' font-size='7.2pt' font-family='DejaVu Sans'>300<\/text>\n <text x='473.75' y='222.22' font-size='7.2pt' font-family='DejaVu Sans'>400<\/text>\n <text x='450.49' y='195.88' font-size='9pt' font-family='DejaVu Sans'>disp<\/text>\n <line x1='450.49' y1='283.16' x2='453.95' y2='283.16' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.37' stroke-linejoin='round' stroke-linecap='butt'/>\n <line x1='450.49' y1='261.68' x2='453.95' y2='261.68' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.37' stroke-linejoin='round' stroke-linecap='butt'/>\n <line x1='450.49' y1='240.2' x2='453.95' y2='240.2' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.37' stroke-linejoin='round' stroke-linecap='butt'/>\n <line x1='450.49' y1='218.72' x2='453.95' y2='218.72' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.37' stroke-linejoin='round' stroke-linecap='butt'/>\n <line x1='464.32' y1='283.16' x2='467.77' y2='283.16' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.37' stroke-linejoin='round' stroke-linecap='butt'/>\n <line x1='464.32' y1='261.68' x2='467.77' y2='261.68' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.37' stroke-linejoin='round' stroke-linecap='butt'/>\n <line x1='464.32' y1='240.2' x2='467.77' y2='240.2' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.37' stroke-linejoin='round' stroke-linecap='butt'/>\n <line x1='464.32' y1='218.72' x2='467.77' y2='218.72' stroke='#FFFFFF' stroke-opacity='1' stroke-width='0.37' stroke-linejoin='round' stroke-linecap='butt'/>\n <\/g>\n <\/g>\n<\/svg>","js":null,"uid":"svg_0c9c18b4_2e68_4579_b86d_088360a75cbf","ratio":1,"settings":{"tooltip":{"css":".tooltip_SVGID_ { padding:5px;background:black;color:white;border-radius:2px;text-align:left; ; position:absolute;pointer-events:none;z-index:999;}","placement":"doc","opacity":0.9,"offx":10,"offy":10,"use_cursor_pos":true,"use_fill":false,"use_stroke":false,"delay_over":200,"delay_out":500},"hover":{"css":".hover_data_SVGID_ { fill:orange;stroke:black;cursor:pointer; }\ntext.hover_data_SVGID_ { stroke:none;fill:orange; }\ncircle.hover_data_SVGID_ { fill:orange;stroke:black; }\nline.hover_data_SVGID_, polyline.hover_data_SVGID_ { fill:none;stroke:orange; }\nrect.hover_data_SVGID_, polygon.hover_data_SVGID_, path.hover_data_SVGID_ { fill:orange;stroke:none; }\nimage.hover_data_SVGID_ { stroke:orange; }","reactive":true,"nearest_distance":null},"hover_inv":{"css":""},"hover_key":{"css":".hover_key_SVGID_ { fill:orange;stroke:black;cursor:pointer; }\ntext.hover_key_SVGID_ { stroke:none;fill:orange; }\ncircle.hover_key_SVGID_ { fill:orange;stroke:black; }\nline.hover_key_SVGID_, polyline.hover_key_SVGID_ { fill:none;stroke:orange; }\nrect.hover_key_SVGID_, polygon.hover_key_SVGID_, path.hover_key_SVGID_ { fill:orange;stroke:none; }\nimage.hover_key_SVGID_ { stroke:orange; }","reactive":true},"hover_theme":{"css":".hover_theme_SVGID_ { fill:orange;stroke:black;cursor:pointer; }\ntext.hover_theme_SVGID_ { stroke:none;fill:orange; }\ncircle.hover_theme_SVGID_ { fill:orange;stroke:black; }\nline.hover_theme_SVGID_, polyline.hover_theme_SVGID_ { fill:none;stroke:orange; }\nrect.hover_theme_SVGID_, polygon.hover_theme_SVGID_, path.hover_theme_SVGID_ { fill:orange;stroke:none; }\nimage.hover_theme_SVGID_ { stroke:orange; }","reactive":true},"select":{"css":".select_data_SVGID_ { fill:red;stroke:black;cursor:pointer; }\ntext.select_data_SVGID_ { stroke:none;fill:red; }\ncircle.select_data_SVGID_ { fill:red;stroke:black; }\nline.select_data_SVGID_, polyline.select_data_SVGID_ { fill:none;stroke:red; }\nrect.select_data_SVGID_, polygon.select_data_SVGID_, path.select_data_SVGID_ { fill:red;stroke:none; }\nimage.select_data_SVGID_ { stroke:red; }","type":"multiple","only_shiny":true,"selected":[]},"select_inv":{"css":""},"select_key":{"css":".select_key_SVGID_ { fill:red;stroke:black;cursor:pointer; }\ntext.select_key_SVGID_ { stroke:none;fill:red; }\ncircle.select_key_SVGID_ { fill:red;stroke:black; }\nline.select_key_SVGID_, polyline.select_key_SVGID_ { fill:none;stroke:red; }\nrect.select_key_SVGID_, polygon.select_key_SVGID_, path.select_key_SVGID_ { fill:red;stroke:none; }\nimage.select_key_SVGID_ { stroke:red; }","type":"single","only_shiny":true,"selected":[]},"select_theme":{"css":".select_theme_SVGID_ { fill:red;stroke:black;cursor:pointer; }\ntext.select_theme_SVGID_ { stroke:none;fill:red; }\ncircle.select_theme_SVGID_ { fill:red;stroke:black; }\nline.select_theme_SVGID_, polyline.select_theme_SVGID_ { fill:none;stroke:red; }\nrect.select_theme_SVGID_, polygon.select_theme_SVGID_, path.select_theme_SVGID_ { fill:red;stroke:none; }\nimage.select_theme_SVGID_ { stroke:red; }","type":"single","only_shiny":true,"selected":[]},"zoom":{"min":1,"max":1,"duration":300},"toolbar":{"position":"topright","pngname":"diagram","tooltips":null,"hidden":[],"delay_over":200,"delay_out":500},"sizing":{"rescale":true,"width":0.3}}},"evals":[],"jsHooks":[]}</script> Other packages for interactive graphics: **ggvis**(see [this link](https://ggvis.rstudio.com/)) and **rCharts** (see [this link](https://ramnathv.github.io/rCharts/)) --- # Short introduction to **Shiny** #### What is shiny ? * **Shiny**: Interactive web page * Examples of shiny app: http://shiny.rstudio.com/gallery/ * More informations [here](https://www.londonr.org/wp-content/uploads/sites/2/presentations/LondonR_-_Workshop-Introduction_to_Shiny_-_Aimee_Gott_-_20150330.pdf) * Create a new shiny App: File - New File - Shiny Web App... * Possibility to create web pages. **RStudio** proposes to host a couple of App for free. See https://www.rstudio.com/products/shiny/shiny-server/ for more informations <img src="Figures/shiny.png" width="50%" height="\textheight" /> --- # The two files .pull-left[ ```r library(shiny) # Define UI for application that draws a histogram ui <- fluidPage( # Application title titlePanel("Old Faithful Geyser Data"), # Sidebar with a slider input for number of bins sidebarLayout( sidebarPanel( sliderInput("bins", "Number of bins:", min = 1, max = 50, value = 30) ), # Show a plot of the generated distribution mainPanel( plotOutput("distPlot") ) ) ) ``` ] .pull-right[ ```r # Define server logic required to draw a histogram server <- function(input, output) { output$distPlot <- renderPlot({ # generate bins based on input$bins from ui.R x <- faithful[, 2] bins <- seq(min(x), max(x), length.out = input$bins + 1) # draw the histogram with the specified number of bins hist(x, breaks = bins, col = 'darkgray', border = 'white') }) } ``` To run the application: ```r shinyApp(ui = ui, server = server) ``` ] --- # Description of the two files * *ui.R*: indicates how the screen should be organized. For example, on the left, we print the title, the ruler, etc. that the user can eventually modify. On the right, we decide to print the graphic. * *server.R*: contains the codes which permits to plot a graphic by using the parameters defined by users in the interface. * When these files are opened in **RStudio**, it is then possible to run the App by clicking on the button Run App. --- # Description of the options #### **ui.R** file * *textInput()*: entering a string, * *numericInput()*: entering a numeric, * *selectInput()*: Create a select list input control, * *sliderInput()*: Slider Input Widget, * *radioButtons()*: Create radio buttons * *fileInput()*: File Upload Control. #### **server.R** file * For plotting a graphic: use *renderPlot()* in **server.R** and *plotOutput()* in **ui.R**. * For printing text: use *renderPrint()* in **server.R** and *textOutput()* in **ui.R**. * For printing data table: use *renderDataTable()* in **server.R** and *dataTableOutput()* in **ui.R**. * For printing images: use *renderImage()* in **server.R** and *imageOutput()* in **ui.R**.